- Home
- /
- Analytics
- /
- Stat Procs
- /
- Generalized linear mixed model - setting and interpreting
- RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Hello all,
I have set up an GLMM model, and I am not 100% sure I have set the right model, while on the other hand struggle to make good interpretation of some of the results.
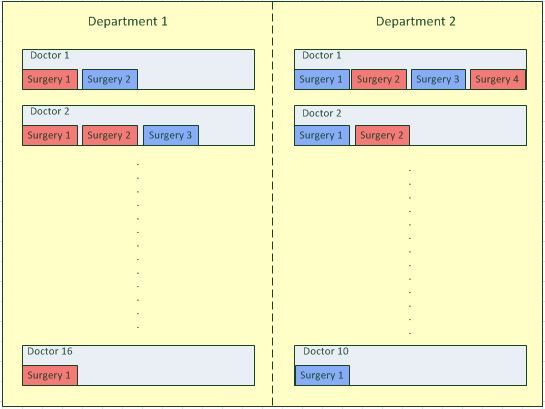
I will try to simplify the problem, and in order to do so, I attach a diagram of the design. I am comparing 2 treatments used in some sort of surgery, a standard treatment, and a new proposed treatment. The outcome of the surgery is success or failure (0/1), i.e., binary.
Two departments were chosen randomly. In each department, doctors were chosen randomly to perform the surgeries. In the 1st department there were 16 doctors chosen, while in the 2nd only 10 (these are big departments).
Each doctor performed between 1 to 4 surgeries. Each surgery was randomly assigned to a treatments (red - standard, blue - new). Each surgery gave an outcome of success or failure.
The diagram is how I see things. Now, I have set the following model:
proc glimmix data = ... method = quad;
class department doctor_id treatment;
model outcome = treatment / dist = binary oddsratio solution;
random int / solution subject = doctor_id(department);
run;
This syntax ran well, with no major errors.
The treatment variable was statistically significant, and the model showed no over dispersion (Pearson Chi-Square / DF = 0.86).
The covariance parameter estimate for the intercept of doctor(department) was 0.326.
When I tried to add another random statement: "random int / solution subject = doctor_id;", I got an error saying that the G matrix is not positive definite. I still got 0.326 for doctor_id(department), and got 4.41E-13 for doctor_id. It didn't look good, so I removed it and went back to the syntax I wrote above (didn't like the error message).
I understand that 0.326 is the estimated variance of the random doctor in department intercept on the logit scale. My main questions thus are:
1. How do I know if the source of the variance is the doctors or departments ? From descriptive view, I am quite confident that it's coming from departments and not doctors, but theoretically, how can I know, if I add another random statement and get an error...by the way, trying to write the syntax different, i.e., adding a random statement for department only and not doctors, doesn't even run.
2. How do I make an interpretation of this results. Let's say that I need to explain this result to someone who doesn't know what logit scale is. How do I do it ? I read somewhere that I can calculate the inter-class correlation (ICC) using this formula: ICC = 0.326 / (0.326+3.29) = 0.0901. It makes sense, the inter-class correlation is not high, meaning that most variance is due to the different treatments and not the random effects (right ?). I am not sure if this formula is correct or not, but with or without a relation to it, how would you report the result of 0.326 so that someone with very basic statistical knowledge will understand you ?
Thank you in advance for any help.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
I don't have time to fully assess your design, but will give you some pointers on your code, and the interpretation of the results. The message about the G matrix is really a note or warning, not an error (usually). In your case, it simply means that your between-doctor variance is 0 (4.4E-13 means 4.4*10^-13, which is basically 0). I would take out the doctor term, as you did, because your results indicate no substantial doctor effect.In fact, it does NOT make sense to include this term. As you describe the design, doctors are nested within departments. There is no real meaning to doctor as a stand-alone term in the model. If you labeled the doctors uniquely across the two departments (for instance, that there cannot be a doctor "1" in each department; perhaps doctors 1-10 in department A and doctors 11-20 in department B), then you could use either
random int / solution subject = doctor_id(department);
or
random int / solution subject = doctor_id;
It would be the same thing in terms of the model for the random effect. This would not be true if there are doctors 1-10 in A and 1-10 in B (for instance). Basically, with nesting, there is an interaction, a consequence of your particular design. With your chosen model, you can't address this (they are tied together).
However, you could fit your model with random effects for department and doctor within department:
random int doctor_id / solution subject = department;
which gives you two random effects (department and doctor nested within department). This can tell you about the effect of department. But with only two departments, the variance estimate for department will be very imprecise. Not sure how much you can interpret it (it may have a large SE). You could get a 0 for one of the variances.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Since you have multiple observations per doctor, I think you should reformat your data file in the y/n format. That is, each record should correspond to a doctor (within a department). Give the number of successful surgeries (y), the number of surgeries for that doctor (n), treatment, department, and anything else. Then the model would be:
proc glimmix data = ... method = quad;
class department doctor_id treatment;
model y/n= treatment / dist = binary oddsratio solution;
random int / solution subject = doctor_id(department);
run;
or
random int doctor_id / subject=department;
if you want to try the hierarchy.
Things are usually better behaved, and the conditional model (what you have by using random effects) has much better properties when n (number of surgeries) is greater than 1. Your coding makes n=1 since an observatin can be either 0 or 1; this can lead to large biases for GLMMs. With these changes, your wish to calculate the intra-cluster correlation may be more appropriate. You are taking the approach that there is an underlying standardized logistic distribution with your correlation formula (where 3.29 is pi^2/3). There are other approaches to this based on different assumptions.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
One slight mistake in my previous message. If you use the y/n format, then use dist=binomial, not dist=binary. If you use the y/n format for the data, then it makes more sense to look at the conditional Pearson chi-squared / df for goodness of fit.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Thank you for your willingness to help !
First I have to apologize, I had in mind to add one more comment, but it slipped from my head, and you commented on this issue. Doctor 1 from department 1, and doctor 1 from department 2, are NOT the same doctors, sorry about the confusion, I realized the confusion after I draw the diagram and forgot to clarify.
I am not sure I fully understand the basic concepts. My head works in a maybe old fashioned way. I understand that the probability of success depends on various reasons (causes of variability if to use anova terms), we have treatment, department, doctor and random error. What I understand from what you wrote, is that doctors has no effect on the probability of success, or nearly no effect. The only effect is the treatment and the department. It makes sense according to graphs I saw. Yet, I am still not sure how to make a proper interpretation of what 0.326 actually mean. Can you please tell me (if there are any) of other methods to evaluate the ICC, which is the only meaningful measure I know of this ?
I didn't expect to encounter any problems with the model statement of my syntax. In my eyes using GLMM was exactly like using Logistic regression, only adding the random effects. When I use logistic regression, in any software, SAS or other, I always use single observations, it's usually the default. I will try your suggestion to see if there will be any differences.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Regarding your last comment, although you can use the data as you coded, you should not do that. With GLMMs (what you have), there can be large biases in fixed effect parameter estimates (not relevant for GLMs, as in PROC LOGISTIC). See SAS for Mixed Models, 2nd edtion (2006), or the new book by Walter Stroup. The larger the n (number of surgeries per doctor), the less the bias when there are random effects. The only time there is a really high bias with a GLMM is with the situation you have (equivalent to n=1). So be careful. Plus, you really should not do a goodness of fit test with the data in your form.
The ICC only makes sense when your data are in the y/n format. The ICC roughly gives the average correlation of binary status of the individuals within the lowest sampling unit. But with your approach, there is one individual within this sampling unit (the success of the surgery). A correlation of one value makes no sense. I will have to look up a citation on other versions of ICC.
I did not say that department or doctor does not have an effect. I gave you code to properly test for a random department effect (you did not test for this, based on your post). But I did say it is hard to test for department when there are only two departments. The results you show indicate that doctor within department has an effect. This is really the same as an interaction of doctor and department. Most of the effect of doctor(department) could be from the department or the doctors. You can consider departments directly, but you can only consider doctors within departments.
One last point for now. You mention random error. For you, doctor(department) is the random error, there is no other random error. You talked about variability, with an ANOVA mindset. I understand what you are trying to get at, but with GLMM, one must move away form ideas like explained variability. THere is a different mindset to fully appreciate the jump from LM to GLM to GLMM. Stroup's new book on generalized linear mixed models explains this in detail.
Good luck.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
I am trying to implement your suggestion to use the y/n format just now, and I seem to be having a problem. Each doctors had 1 or more surgeries, but sometimes with different treatments. How should I aggregate the data ? Should I make distinct rows for the same doctor, one for each treatment ?
Example: If doctor #1 had 4 surgeries, 2 with treatment A and 2 with treatment B, and in treatment A both were a success, while in treatment B only 1, how should I aggregate this data to be read by your suggested model ?
ANOVA, or Analysis Of Variance, is used to compare the averages or means of two or more populations to better understand how they differ. Watch this tutorial for more.
Find more tutorials on the SAS Users YouTube channel.


