- Home
- /
- Programming
- /
- Graphics
- /
- Re: Errant Graph when ploting predicted values from proc nlin
- RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Hello,
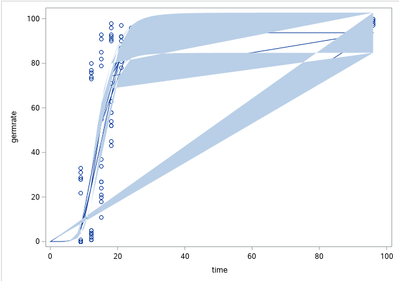
I'm currently working on a SAS Programm to fit my data of a germinating fungus to a given logistic function. This works fine for now, as the estimated values fit well to the data and predicted values based on that function seem acceptable. But when I try to plot this data the resulting graph looks like this:
Does anyone has any suggestions regarding this problem? I'm using SAS University Edition / SAS Studio 3.8
Thanks for your help!
The code used to create this graph is as follows:
data fungi;
input time germrate;
datalines;
9 0
9 0
9 0
9 0
9 0
9 0
9 0
9 0
12 1
12 1
12 1
9 1
12 2
12 3
12 4
12 4
12 5
12 5
15 11
15 18
15 20
15 21
9 22
15 24
15 27
15 27
9 28
9 28
9 29
9 31
9 33
15 34
15 37
18 43
18 45
18 52
18 52
18 54
15 55
18 58
18 58
18 63
18 71
21 72
12 73
21 74
12 74
12 76
12 76
12 77
21 78
15 79
12 80
21 81
15 82
21 83
21 84
21 84
24 85
15 85
24 86
21 86
24 87
21 87
21 87
24 88
24 90
24 90
24 90
21 90
18 90
18 91
15 91
24 92
24 92
21 92
21 92
21 92
18 92
24 93
24 93
18 93
15 93
24 94
24 94
24 95
24 96
18 96
21 97
18 98
96 99
96 100
96 99
96 98
96 97
96 100
;
proc nlin data=fungi;
parms Pmax=98 teta=15 ;d=6 ;
model germrate = Pmax*(1-(1/(1+((time/teta)**d))));
run;
proc nlin data=fungi method=newton;
parameters Pmax=98 teta=15 ;d=6 ;
model germrate = Pmax*(1-(1/(1+((time/teta)**d))));
output out=nlinout predicted=pred l95m=l95mean u95m=u95mean l95=l95ind u95=u95ind;
run;
proc print data=nlinout;
run;
data filler;
do time = 0 to 96 by 2;
predict=1;
output;
end;
run;
data fitthis;
set fungi filler;
run;
proc nlin data=fitthis method=newton;
parameters Pmax=98 teta=15 ;d=6 ;
model germrate = Pmax*(1-(1/(1+((time/teta)**d))));
output out=nlinout predicted=pred l95m=l95mean u95m=u95mean;
run;
proc print data=nlinout(where=(predict=1)); run;
proc sgplot data=nlinout;
scatter x=time y=germrate;
series x=time y=pred;
band upper=u95mean lower=l95mean x=time ;
run;
Accepted Solutions
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Hi @RobinN , you just need to sort your graph by time before you do the plot. Please see the example below.
data fungi;
input time germrate;
datalines;
9 0
9 0
9 0
9 0
9 0
9 0
9 0
9 0
12 1
12 1
12 1
9 1
12 2
12 3
12 4
12 4
12 5
12 5
15 11
15 18
15 20
15 21
9 22
15 24
15 27
15 27
9 28
9 28
9 29
9 31
9 33
15 34
15 37
18 43
18 45
18 52
18 52
18 54
15 55
18 58
18 58
18 63
18 71
21 72
12 73
21 74
12 74
12 76
12 76
12 77
21 78
15 79
12 80
21 81
15 82
21 83
21 84
21 84
24 85
15 85
24 86
21 86
24 87
21 87
21 87
24 88
24 90
24 90
24 90
21 90
18 90
18 91
15 91
24 92
24 92
21 92
21 92
21 92
18 92
24 93
24 93
18 93
15 93
24 94
24 94
24 95
24 96
18 96
21 97
18 98
96 99
96 100
96 99
96 98
96 97
96 100
;
proc nlin data=fungi;
parms Pmax=98 teta=15 ;d=6 ;
model germrate = Pmax*(1-(1/(1+((time/teta)**d))));
run;
proc nlin data=fungi method=newton;
parameters Pmax=98 teta=15 ;d=6 ;
model germrate = Pmax*(1-(1/(1+((time/teta)**d))));
output out=nlinout predicted=pred l95m=l95mean u95m=u95mean l95=l95ind u95=u95ind;
run;
proc print data=nlinout;
run;
data filler;
do time = 0 to 96 by 2;
predict=1;
output;
end;
run;
data fitthis;
set fungi filler;
run;
proc nlin data=fitthis method=newton;
parameters Pmax=98 teta=15 ;d=6 ;
model germrate = Pmax*(1-(1/(1+((time/teta)**d))));
output out=nlinout predicted=pred l95m=l95mean u95m=u95mean;
run;
proc print data=nlinout(where=(predict=1)); run;
proc sort data = nlinout;
by time;
run;
proc sgplot data=nlinout;
scatter x=time y=germrate;
series x=time y=pred;
band upper=u95mean lower=l95mean x=time ;
run;- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Hi @RobinN , you just need to sort your graph by time before you do the plot. Please see the example below.
data fungi;
input time germrate;
datalines;
9 0
9 0
9 0
9 0
9 0
9 0
9 0
9 0
12 1
12 1
12 1
9 1
12 2
12 3
12 4
12 4
12 5
12 5
15 11
15 18
15 20
15 21
9 22
15 24
15 27
15 27
9 28
9 28
9 29
9 31
9 33
15 34
15 37
18 43
18 45
18 52
18 52
18 54
15 55
18 58
18 58
18 63
18 71
21 72
12 73
21 74
12 74
12 76
12 76
12 77
21 78
15 79
12 80
21 81
15 82
21 83
21 84
21 84
24 85
15 85
24 86
21 86
24 87
21 87
21 87
24 88
24 90
24 90
24 90
21 90
18 90
18 91
15 91
24 92
24 92
21 92
21 92
21 92
18 92
24 93
24 93
18 93
15 93
24 94
24 94
24 95
24 96
18 96
21 97
18 98
96 99
96 100
96 99
96 98
96 97
96 100
;
proc nlin data=fungi;
parms Pmax=98 teta=15 ;d=6 ;
model germrate = Pmax*(1-(1/(1+((time/teta)**d))));
run;
proc nlin data=fungi method=newton;
parameters Pmax=98 teta=15 ;d=6 ;
model germrate = Pmax*(1-(1/(1+((time/teta)**d))));
output out=nlinout predicted=pred l95m=l95mean u95m=u95mean l95=l95ind u95=u95ind;
run;
proc print data=nlinout;
run;
data filler;
do time = 0 to 96 by 2;
predict=1;
output;
end;
run;
data fitthis;
set fungi filler;
run;
proc nlin data=fitthis method=newton;
parameters Pmax=98 teta=15 ;d=6 ;
model germrate = Pmax*(1-(1/(1+((time/teta)**d))));
output out=nlinout predicted=pred l95m=l95mean u95m=u95mean;
run;
proc print data=nlinout(where=(predict=1)); run;
proc sort data = nlinout;
by time;
run;
proc sgplot data=nlinout;
scatter x=time y=germrate;
series x=time y=pred;
band upper=u95mean lower=l95mean x=time ;
run;- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
April 27 – 30 | Gaylord Texan | Grapevine, Texas
Registration is open
Walk in ready to learn. Walk out ready to deliver. This is the data and AI conference you can't afford to miss.
Register now and save with the early bird rate—just $795!
Learn how use the CAT functions in SAS to join values from multiple variables into a single value.
Find more tutorials on the SAS Users YouTube channel.
SAS Training: Just a Click Away
Ready to level-up your skills? Choose your own adventure.