- Home
- /
- Analytics
- /
- Stat Procs
- /
- Doubts about proc genmod
- RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Hi,
I am trying to generate a model to predict the number of absences of a school alumn. My inputs var are: math and literature marks of the alumns and their sex. The number of absences vary from 0 to 12. I am using a GLM model with 'proc genmod'
proc genmod data = test;
class sexo;
model diasau = nota_mates nota_lengua sexo / dist=Poisson;
store out=modelo_glm;
run;I have some doubts:
- How can I select the apropiate distibution: Poisson...?
- Do I have to do some log transformation?
I am not an expert in GLM models, but I know that I have to review the model deviance vs degree of freedoms:
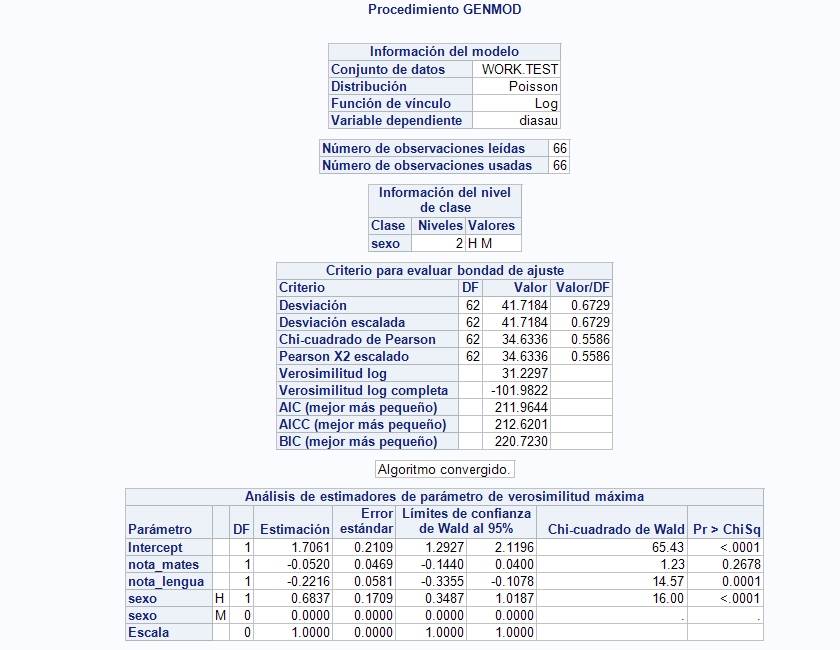
Thats what I get:
Any advice will be greatly appreciated.
Thanks in advance?
Accepted Solutions
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
A count response is most typically modeled using the Poisson or the negative binomial distribution. The negative binomial distribution is used when the Poisson model shows evidence of overdispersion as discussed in this note. Since your "Value/DF" values are all less than 1, there doesn't seem to be any evidence of overdispersion. When fitting either of those distributions, the log link function is the default, so that the model is log(mean) = intercept + b1*x1 + b2*x2 + ... . So, you should not apply any transformation to your response.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
A count response is most typically modeled using the Poisson or the negative binomial distribution. The negative binomial distribution is used when the Poisson model shows evidence of overdispersion as discussed in this note. Since your "Value/DF" values are all less than 1, there doesn't seem to be any evidence of overdispersion. When fitting either of those distributions, the log link function is the default, so that the model is log(mean) = intercept + b1*x1 + b2*x2 + ... . So, you should not apply any transformation to your response.
ANOVA, or Analysis Of Variance, is used to compare the averages or means of two or more populations to better understand how they differ. Watch this tutorial for more.
Find more tutorials on the SAS Users YouTube channel.