- Home
- /
- Programming
- /
- Graphics
- /
- Pie-like map colored by slice with frequency heat
- RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Greetings,
I would like to create a map that looks like a sliced pie face (8 pieces with cut-lines at specified positions) and each pie slice colored to represent a count. I do not want a pie-chart in which pie slice area represent relative count information. Rather, the pie face represents a sampling grid (a map) in which I want color to represent frequency of sampling within each site. I've not done this before and could use your help to point me in the right direction with procedure selection and application.
Thank you,
Dave
Accepted Solutions
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Das;
A bar chart (V or H) is much easier to understand than a spider/radar chart. (http://www.perceptualedge.com/articles/dmreview/radar_graphs.pdf).
Take a look at what I've included below. Notice too, that a value zero can be displayed and, for goat at position 5 is very obvious!
data bx_position ;
input specie $ position count ;
datalines;
goat 1 1
goat 2 5
goat 3 65
goat 4 27
goat 5 0
goat 6 18
goat 7 37
goat 8 2
sheep 1 1
sheep 2 6
sheep 3 78
sheep 4 32
sheep 5 2
sheep 6 14
sheep 7 45
sheep 8 1
;
run;
proc gchart;
hbar specie/group=position sumvar=count patterid=midpoint;
run;
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Can you provide an example drawing or other graphic of what you want? I'm not sure I understand and since this relates to pictures the 1:1000 ratio needs some help.
I suspect that an annotate dataset with several PIE statements would generate the pie bits and the colors can be specified at creation. Then additional statements, LABEL perhaps could place text.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Here are two possibilities. I suspect the first is the simplest and would suffice. The numbers are the frequency on which color choice and/or shading would depend.
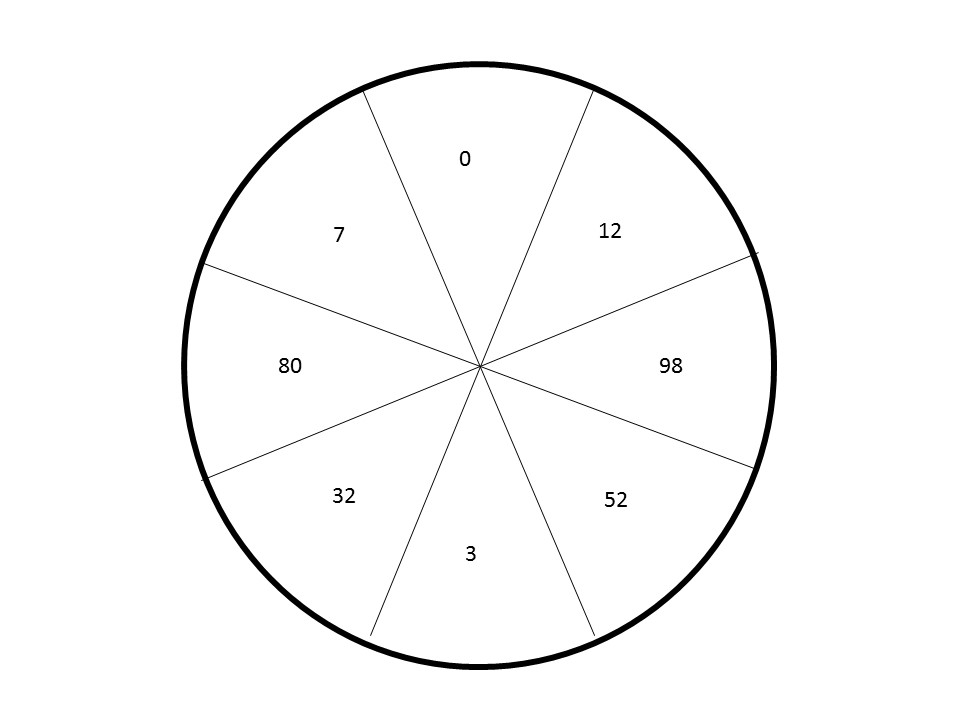
It is actually supposed to be a map of biopsy regions of a tubular organ. So the following would be a more optimal graphic.

Thanks for all suggestions.
Dave
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Something like this may give you enough to get started:
%annomac;
data work.pie;
%dclanno;
hsys='3';xsys='3';ysys='3';
%slice(50,50,0,45,20,red,PS,both);
%slice(50,50,.,45,20,blue,PS,both);
%slice(50,50,.,45,20,orange,PS,both);
%slice(50,50,.,45,20,green,PS,both);
%slice(50,50,.,45,20,pink,PS,both);
%slice(50,50,.,45,20,lightblue,PS,both);
%slice(50,50,.,45,20,gold,PS,both);
%slice(50,50,.,45,20,lightgreen,PS,both);
run;
proc gslide anno=work.pie;
run;
quit;
If you have add a slice of 360 degrees with a rad of 10 or so and color white at the end of the data I think you'll get the tube.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Thank you. That is very helpful. I was struggling to even get myself to go down this path. I think I can modify this to satisfy my needs. I also started looking at radar/polar graphs as another means to my end.
Dave
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
I'm liking the representation using the proc gradar. Here is an image closing in on something acceptable:

Here the vertices correspond to the biopsy position of the tubular organ with position "1" at 12 o'clock high. The problems are that this procedure doesn't seem to map a zero frequency and so I've had to change these to "1" in the count in order for the spoke position to show up in the graph. Suggestions still appreciated. I realize I've gotten away from my original intent on how to graph but still appreciative of any advice.
Here is the code:
data bx_position ;
input specie $ position count ;
datalines;
goat 1 1
goat 2 5
goat 3 65
goat 4 27
goat 5 1
goat 6 18
goat 7 37
goat 8 2
sheep 1 1
sheep 2 6
sheep 3 78
sheep 4 32
sheep 5 2
sheep 6 14
sheep 7 45
sheep 8 1
;
run;
proc gradar data=bx_position;
chart position /
freq=COUNT
overlay=specie
startype=wedge
lstar=1 1 1
cstar=(red blue black)
;
run;
quit;
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Das;
A bar chart (V or H) is much easier to understand than a spider/radar chart. (http://www.perceptualedge.com/articles/dmreview/radar_graphs.pdf).
Take a look at what I've included below. Notice too, that a value zero can be displayed and, for goat at position 5 is very obvious!
data bx_position ;
input specie $ position count ;
datalines;
goat 1 1
goat 2 5
goat 3 65
goat 4 27
goat 5 0
goat 6 18
goat 7 37
goat 8 2
sheep 1 1
sheep 2 6
sheep 3 78
sheep 4 32
sheep 5 2
sheep 6 14
sheep 7 45
sheep 8 1
;
run;
proc gchart;
hbar specie/group=position sumvar=count patterid=midpoint;
run;
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Thank you for the input. I remain torn between the simple bar chart versus the original intent to visually map the biopsy frequency to an intuitive cartoon-like map of the tubular organ biopsied. I note that gradar can produce bar-like graphs by choosing the "spoke" formatted chart (see first two images below). However, I think your suggestion but as a vertical bar, in which the order from left-to-right might be enough intuitive orientation, especially if I change the position numbers to their corresponding positions on a clock face (see third image). That'll make left to right sideness intuitive and only lose the intuitiveness of the up versus down positions. But I think I most agree that some form of a bar graph will be the most easily created AND understood. Just got to get the axis straightened out.
Thank you for the suggestion and code example. Very helpful!
Dave



Learn how use the CAT functions in SAS to join values from multiple variables into a single value.
Find more tutorials on the SAS Users YouTube channel.
SAS Training: Just a Click Away
Ready to level-up your skills? Choose your own adventure.





