- Home
- /
- Programming
- /
- Graphics
- /
- Re: NEED HELP about Swimmer Plot
- RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Hi, I'm having trouble when I output Swimmer plot.
First question, I'd like to understand how to adjust the size of y-axis value or the line-width (x-axis is Week and y-axis is Subject No.) because it would not show all subject no. when the number bars is too many.
Second, there's an uneven color of each bar under option transparency=0.X when one subject has two or more events. I'd like to know how to resolve that issue if I still have to adjust the transparency of bars.
This is my first time to share questions here, please feel free to give me advice when you can.
proc sgplot data=final nocycleattrs;
highlow y=ptno low=low high=line_end / group=stage attrid=A type=bar fill nooutline transparency=0.5
name='stage' barwidth=0.3;
highlow y=ptno low=crstart1 high=crend1 / group=Duration lineattrs=(thickness=2 pattern=solid)
lineattrs=(color=red) name='Duration' nomissinggroup attrid=Duration;
highlow y=ptno low=crstart2 high=crend2 / group=Duration lineattrs=(thickness=2 pattern=solid)
lineattrs=(color=blue) name='Duration' nomissinggroup attrid=Duration;
highlow y=ptno low=crstart3 high=crend3 / group=Duration lineattrs=(thickness=2 pattern=solid)
lineattrs=(color=black) name='Duration' nomissinggroup attrid=Duration;
scatter X=crstart1 Y=ptno /markerattrs=(symbol=trianglefilled size=12 color=red) name='a' legendlabel='Mild Constipation Start';
scatter X=crend1 Y=ptno /markerattrs=(symbol=circlefilled size=12 color=red) name='b' legendlabel='Mild Constipation End';
scatter X=crstart2 Y=ptno /markerattrs=(symbol=trianglefilled size=12 color=blue) name='c' legendlabel='Moderate Constipation Start';
scatter X=crend2 Y=ptno /markerattrs=(symbol=circlefilled size=12 color=blue) name='d' legendlabel='Moderate Constipation End';
scatter X=crstart3 Y=ptno /markerattrs=(symbol=trianglefilled size=12 color=black) name='e' legendlabel='Severe Constipation Start';
scatter X=crend3 Y=ptno /markerattrs=(symbol=circlefilled size=12 color=black) name='f' legendlabel='Severe Constipation End';
yaxis type=discrete display=(nolabel noticks);
xaxis type=linear label="Week" values=(0 to 12 by 1);
keylegend 'stage' / title='Treatment Group';
keylegend 'a' 'b' 'c' 'd' 'e' 'f'/ noborder location=inside position=bottomright across=1;
run;
quit;Accepted Solutions
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Hi John,
There are at least two solutions to help you to view each subject id.
One of them is to change the dimensions of the image, so that it is portrait instead of landscape. This will give you more space on the yaxis I can be done by simply changing the options within ods graphics (I modified Sanjay's earlier example):
ods graphics / reset imagename='Swimmer_portrait' height=6.67in width = 5in;
Another option, as Sanjay also said is to get rid of the labels on the y-axis and add the subjects labels at the end of the bar chart.
The following code, can help you achieve that: Basically the dataset has been updated to include the position where the subject labels should go, and then this label has been added to the end of the barchart using a new highlow statement - just because some of the start values, where after the end values.
* Make labels on bars;
data test2;
set test;
max_end = max(line_end, end1, end2, end3, start1, start2, start3);
label_x_axis = max_end + 0.1;
run;
ods listing gpath='C:\Users\yex7977\Communities\Image' image_dpi=200;
ods graphics / reset imagename='Swimmer_label';
title h=1 'Figure 1 : Summary Horizontal Bar Chart of Severity';
proc sgplot data=test2 nocycleattrs;
* label;
highlow y=No_ low=low high=label_x_axis / attrid=A type=bar lineattrs=(color=white) nofill outline transparency=0.5
name='drug' highlabel=No_;
highlow y=No_ low=low high=line_end / group=drug attrid=A type=bar fill nooutline transparency=0.5
name='drug';
highlow y=No_ low=start1 high=end1 / group=Duration lineattrs=(thickness=2 pattern=solid)
lineattrs=(color=red) name='Duration' nomissinggroup attrid=Duration;
highlow y=No_ low=start2 high=end2 / group=Duration lineattrs=(thickness=2 pattern=solid)
lineattrs=(color=blue) name='Duration' nomissinggroup attrid=Duration;
highlow y=No_ low=start3 high=end3 / group=Duration lineattrs=(thickness=2 pattern=solid)
lineattrs=(color=black) name='Duration' nomissinggroup attrid=Duration;
scatter X=start1 Y=No_ /markerattrs=(symbol=trianglefilled size=8 color=red) name='a' legendlabel='Mild Start';
scatter X=end1 Y=No_ /markerattrs=(symbol=circlefilled size=8 color=red) name='b' legendlabel='Mild End';
scatter X=start2 Y=No_ /markerattrs=(symbol=trianglefilled size=8 color=blue) name='c' legendlabel='Moderate Start';
scatter X=end2 Y=No_ /markerattrs=(symbol=circlefilled size=8 color=blue) name='d' legendlabel='Moderate End';
scatter X=start3 Y=No_ /markerattrs=(symbol=trianglefilled size=8 color=black) name='e' legendlabel='Severe Start';
scatter X=end3 Y=No_ /markerattrs=(symbol=circlefilled size=8 color=black) name='f' legendlabel='Severe End';
yaxis type=discrete display=NONE fitpolicy=none labelpos=top;
xaxis type=linear label="Week" values=(0 to 12 by 1);
keylegend 'drug' / title='Treatment Group';
keylegend 'a' 'b' 'c' 'd' 'e' 'f'/ noborder location=inside position=bottomright across=1;
run;

- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
It is easier to help if you share everything needed to run your program, including data. What release of SAS are you running?
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
DATA:
| No. | Start_date | End_date | Baseline_date | GP | Severity | line_end | start1 | end1 | start2 | end2 | start3 | end3 | low | Duration |
| 101 | 2016/8/26 | 2016/9/3 | 2016/8/19 | A | Moderate | 3.1 | . | . | 2 | 3.1 | . | . | 0 | Duration |
| 102 | 2016/8/31 | 2016/9/13 | 2016/8/19 | B | Mild | 4.6 | 2.7 | 4.6 | . | . | . | . | 0 | Duration |
| 103 | 2016/8/26 | 2016/9/16 | 2016/8/19 | A | Mild | 5 | 2 | 5 | . | . | . | . | 0 | Duration |
| 103 | 2016/8/31 | 2016/8/31 | 2016/8/19 | A | Severe | 2.7 | . | . | . | . | 2.7 | 2.7 | 0 | Duration |
| 103 | 2016/8/25 | 2016/8/30 | 2016/8/19 | A | Severe | 3.1 | . | . | . | . | 1.9 | 2.6 | 0 | Duration |
| 106 | 2016/8/31 | 2016/9/2 | 2016/8/19 | A | Severe | 3.1 | . | . | . | . | 2.7 | 3 | 0 | Duration |
| 107 | 2016/9/3 | 2016/9/3 | 2016/8/19 | A | Severe | 3.1 | . | . | . | . | 3.1 | 3.1 | 0 | Duration |
| 108 | 2016/8/21 | 2016/8/26 | 2016/8/19 | A | Moderate | 3.9 | . | . | 1.3 | 2 | . | . | 0 | Duration |
| 109 | 2016/9/3 | 2016/9/8 | 2016/8/19 | A | Moderate | 3.9 | . | . | 3.1 | 3.9 | . | . | 0 | Duration |
| 110 | 2016/8/30 | 2016/10/13 | 2016/8/19 | C | Moderate | 8.9 | . | . | 2.6 | 8.9 | . | . | 0 | Duration |
| 111 | 2016/9/12 | 2016/9/12 | 2016/8/19 | B | Mild | 4.7 | 4.4 | 4.4 | . | . | . | . | 0 | Duration |
| 112 | 2016/9/14 | 2016/9/14 | 2016/8/19 | B | Mild | 4.7 | 4.7 | 4.7 | . | . | . | . | 0 | Duration |
| 113 | 2016/8/30 | 2016/9/3 | 2016/8/19 | C | Severe | 3.1 | . | . | . | . | 2.6 | 3.1 | 0 | Duration |
| 113 | 2016/9/5 | 2016/9/26 | 2016/8/19 | B | Mild | 6.4 | 3.4 | 6.4 | . | . | . | . | 0 | Duration |
| 113 | 2016/8/28 | 2016/9/6 | 2016/8/19 | A | Severe | 3.6 | . | . | . | . | 2.3 | 3.6 | 0 | Duration |
| 116 | 2016/8/29 | 2016/9/30 | 2016/8/19 | A | Mild | 7 | 2.4 | 7 | . | . | . | . | 0 | Duration |
| 117 | 2016/8/31 | 2016/9/9 | 2016/8/19 | A | Moderate | 4 | . | . | 2.7 | 4 | . | . | 0 | Duration |
| 118 | 2016/8/19 | 2016/9/1 | 2016/8/19 | A | Mild | 2.9 | 1 | 2.9 | . | . | . | . | 0 | Duration |
| 119 | 2016/9/6 | 2016/10/13 | 2016/8/19 | A | Moderate | 8.9 | . | . | 3.6 | 8.9 | . | . | 0 | Duration |
| 120 | 2016/8/29 | 2016/9/2 | 2016/8/19 | A | Moderate | 8.9 | . | . | 2.4 | 3 | . | . | 0 | Duration |
| 121 | 2016/9/1 | 2016/10/8 | 2016/8/19 | A | Mild | 8.1 | 2.9 | 8.1 | . | . | . | . | 0 | Duration |
| 122 | 2016/8/29 | 2016/9/3 | 2016/8/19 | B | Moderate | 3.1 | . | . | 2.4 | 3.1 | . | . | 0 | Duration |
| 123 | 2016/9/1 | 2016/9/7 | 2016/8/19 | A | Moderate | 3.7 | . | . | 2.9 | 3.7 | . | . | 0 | Duration |
| 124 | 2016/8/30 | 2016/9/2 | 2016/8/19 | B | Severe | 3 | . | . | . | . | 2.6 | 3 | 0 | Duration |
| 125 | 2016/8/30 | 2016/9/6 | 2016/8/19 | A | Moderate | 3.6 | . | . | 2.6 | 3.6 | . | . | 0 | Duration |
| 126 | 2016/8/30 | 2016/8/30 | 2016/8/19 | B | Mild | 3.1 | 2.6 | 2.6 | . | . | . | . | 0 | Duration |
| 127 | 2016/8/31 | 2016/9/3 | 2016/8/19 | B | Mild | 3.1 | 2.7 | 3.1 | . | . | . | . | 0 | Duration |
| 128 | 2016/9/6 | 2016/9/8 | 2016/8/19 | B | Moderate | 3.9 | . | . | 3.6 | 3.9 | . | . | 0 | Duration |
| 129 | 2016/8/19 | 2016/9/2 | 2016/8/19 | B | Moderate | 3.9 | . | . | 1 | 3 | . | . | 0 | Duration |
| 130 | 2016/8/29 | 2016/10/14 | 2016/8/19 | B | Moderate | 9 | . | . | 2.4 | 9 | . | . | 0 | Duration |
| 131 | 2016/9/4 | 2016/9/3 | 2016/8/19 | B | Severe | 3.1 | . | . | . | . | 3.3 | 3.1 | 0 | Duration |
| 132 | 2016/8/29 | 2016/9/4 | 2016/8/19 | A | Moderate | 3.3 | . | . | 2.4 | 3.3 | . | . | 0 | Duration |
CODE:
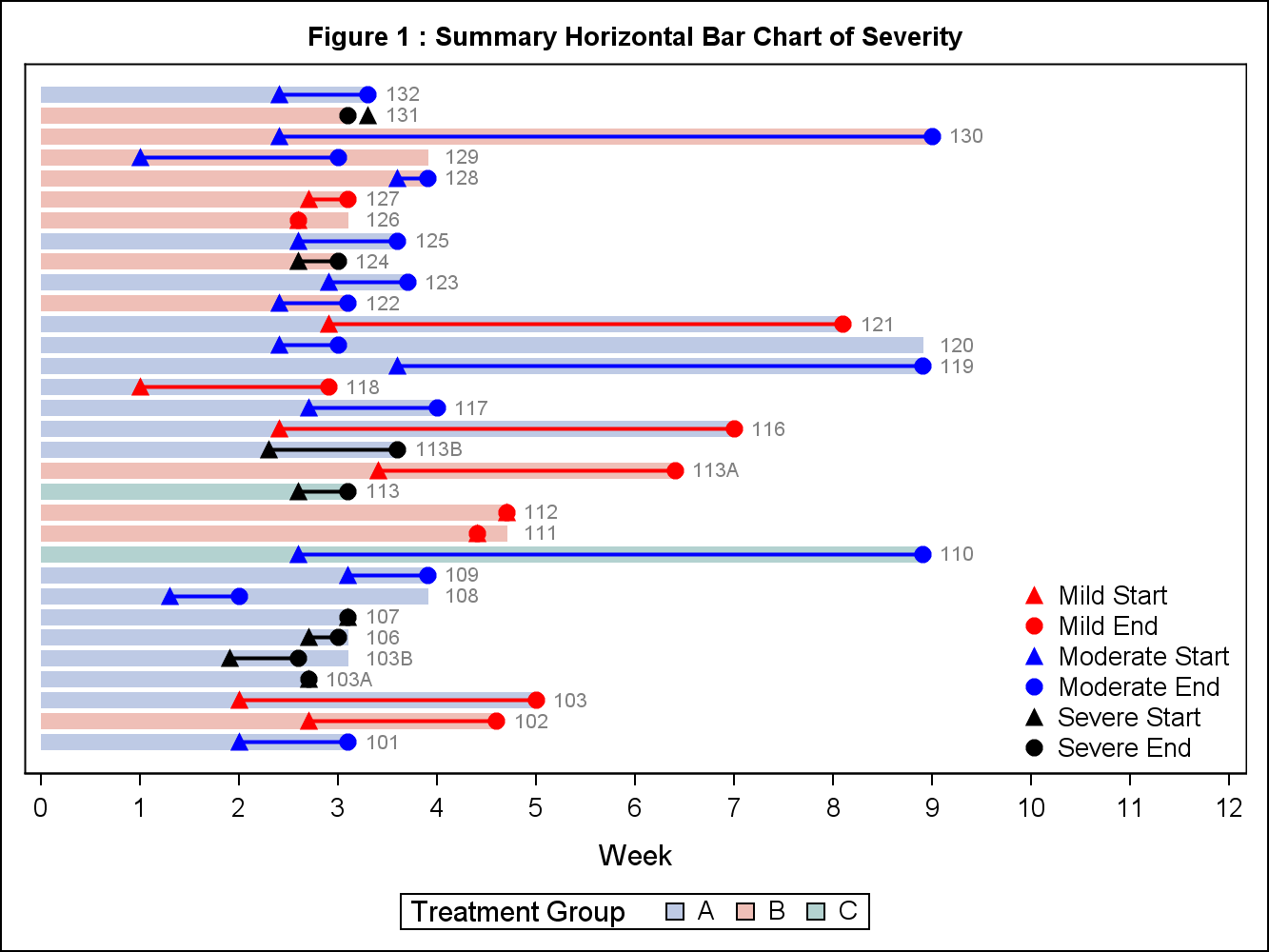
title h=1 'Figure 1 : Summary Horizontal Bar Chart of Severity';
proc sgplot data=test nocycleattrs;
highlow y=No_ low=low high=line_end / group=drug attrid=A type=bar fill nooutline transparency=0.5
name='drug';
highlow y=No_ low=start1 high=end1 / group=Duration lineattrs=(thickness=2 pattern=solid)
lineattrs=(color=red) name='Duration' nomissinggroup attrid=Duration;
highlow y=No_ low=start2 high=end2 / group=Duration lineattrs=(thickness=2 pattern=solid)
lineattrs=(color=blue) name='Duration' nomissinggroup attrid=Duration;
highlow y=No_ low=start3 high=end3 / group=Duration lineattrs=(thickness=2 pattern=solid)
lineattrs=(color=black) name='Duration' nomissinggroup attrid=Duration;
scatter X=start1 Y=No_ /markerattrs=(symbol=trianglefilled size=8 color=red) name='a' legendlabel='Mild Start';
scatter X=end1 Y=No_ /markerattrs=(symbol=circlefilled size=8 color=red) name='b' legendlabel='Mild End';
scatter X=start2 Y=No_ /markerattrs=(symbol=trianglefilled size=8 color=blue) name='c' legendlabel='Moderate Start';
scatter X=end2 Y=No_ /markerattrs=(symbol=circlefilled size=8 color=blue) name='d' legendlabel='Moderate End';
scatter X=start3 Y=No_ /markerattrs=(symbol=trianglefilled size=8 color=black) name='e' legendlabel='Severe Start';
scatter X=end3 Y=No_ /markerattrs=(symbol=circlefilled size=8 color=black) name='f' legendlabel='Severe End';
yaxis type=discrete display=(nolabel noticks);
xaxis type=linear label="Week" values=(0 to 12 by 1);
keylegend 'drug' / title='Treatment Group';
keylegend 'a' 'b' 'c' 'd' 'e' 'f'/ noborder location=inside position=bottomright across=1;
run;
quit;Plot:
Another question, how can I set the wording "Number" in the upper left? Thank you so much!
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
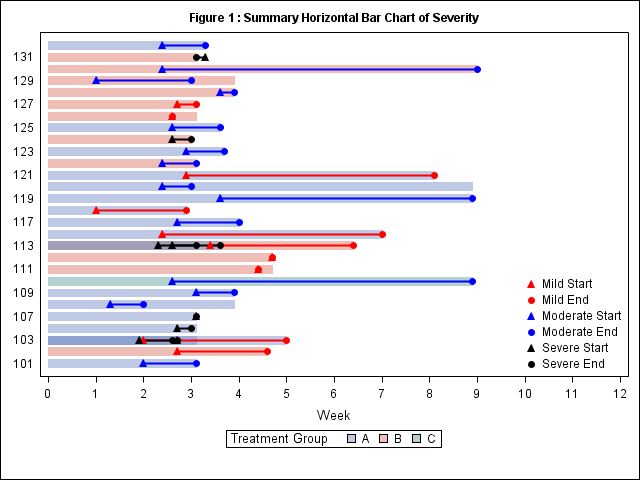
I got your program running. In future it is better to attach SAS code for data too.
All your start values are zero. So, if a subject has multiple events, they will overlap. Either they need different start value, or you need to separate them into separate ids (113, 113A, 113B)..
You can use LabelPos=Top for putting y-axis label on top.
data test;
informat start_date end_date baseline_date anydtdte10.;
format start_date end_date baseline_date date9.;
label No_='Subject';
input No_ $ Start_date End_date Baseline_date Drug $ Severity $ line_end start1 end1 start2 end2 start3 end3 low Duration $;
datalines;
101 2016/8/26 2016/9/3 2016/8/19 A Moderate 3.1 . . 2 3.1 . . 0 Duration
102 2016/8/31 2016/9/13 2016/8/19 B Mild 4.6 2.7 4.6 . . . . 0 Duration
103 2016/8/26 2016/9/16 2016/8/19 A Mild 5 2 5 . . . . 0 Duration
103A 2016/8/31 2016/8/31 2016/8/19 A Severe 2.7 . . . . 2.7 2.7 0 Duration
103B 2016/8/25 2016/8/30 2016/8/19 A Severe 3.1 . . . . 1.9 2.6 0 Duration
106 2016/8/31 2016/9/2 2016/8/19 A Severe 3.1 . . . . 2.7 3 0 Duration
107 2016/9/3 2016/9/3 2016/8/19 A Severe 3.1 . . . . 3.1 3.1 0 Duration
108 2016/8/21 2016/8/26 2016/8/19 A Moderate 3.9 . . 1.3 2 . . 0 Duration
109 2016/9/3 2016/9/8 2016/8/19 A Moderate 3.9 . . 3.1 3.9 . . 0 Duration
110 2016/8/30 2016/10/13 2016/8/19 C Moderate 8.9 . . 2.6 8.9 . . 0 Duration
111 2016/9/12 2016/9/12 2016/8/19 B Mild 4.7 4.4 4.4 . . . . 0 Duration
112 2016/9/14 2016/9/14 2016/8/19 B Mild 4.7 4.7 4.7 . . . . 0 Duration
113 2016/8/30 2016/9/3 2016/8/19 C Severe 3.1 . . . . 2.6 3.1 0 Duration
113A 2016/9/5 2016/9/26 2016/8/19 B Mild 6.4 3.4 6.4 . . . . 0 Duration
113B 2016/8/28 2016/9/6 2016/8/19 A Severe 3.6 . . . . 2.3 3.6 0 Duration
116 2016/8/29 2016/9/30 2016/8/19 A Mild 7 2.4 7 . . . . 0 Duration
117 2016/8/31 2016/9/9 2016/8/19 A Moderate 4 . . 2.7 4 . . 0 Duration
118 2016/8/19 2016/9/1 2016/8/19 A Mild 2.9 1 2.9 . . . . 0 Duration
119 2016/9/6 2016/10/13 2016/8/19 A Moderate 8.9 . . 3.6 8.9 . . 0 Duration
120 2016/8/29 2016/9/2 2016/8/19 A Moderate 8.9 . . 2.4 3 . . 0 Duration
121 2016/9/1 2016/10/8 2016/8/19 A Mild 8.1 2.9 8.1 . . . . 0 Duration
122 2016/8/29 2016/9/3 2016/8/19 B Moderate 3.1 . . 2.4 3.1 . . 0 Duration
123 2016/9/1 2016/9/7 2016/8/19 A Moderate 3.7 . . 2.9 3.7 . . 0 Duration
124 2016/8/30 2016/9/2 2016/8/19 B Severe 3 . . . . 2.6 3 0 Duration
125 2016/8/30 2016/9/6 2016/8/19 A Moderate 3.6 . . 2.6 3.6 . . 0 Duration
126 2016/8/30 2016/8/30 2016/8/19 B Mild 3.1 2.6 2.6 . . . . 0 Duration
127 2016/8/31 2016/9/3 2016/8/19 B Mild 3.1 2.7 3.1 . . . . 0 Duration
128 2016/9/6 2016/9/8 2016/8/19 B Moderate 3.9 . . 3.6 3.9 . . 0 Duration
129 2016/8/19 2016/9/2 2016/8/19 B Moderate 3.9 . . 1 3 . . 0 Duration
130 2016/8/29 2016/10/14 2016/8/19 B Moderate 9 . . 2.4 9 . . 0 Duration
131 2016/9/4 2016/9/3 2016/8/19 B Severe 3.1 . . . . 3.3 3.1 0 Duration
132 2016/8/29 2016/9/4 2016/8/19 A Moderate 3.3 . . 2.4 3.3 . . 0 Duration
;
run;
/*proc print;run;*/
ods listing gpath='C:\Work\SASUser\Communities\Image' image_dpi=200;
ods graphics / reset imagename='Swimmer';
title h=1 'Figure 1 : Summary Horizontal Bar Chart of Severity';
proc sgplot data=test nocycleattrs;
highlow y=No_ low=low high=line_end / group=drug attrid=A type=bar fill nooutline transparency=0.5
name='drug';
highlow y=No_ low=start1 high=end1 / group=Duration lineattrs=(thickness=2 pattern=solid)
lineattrs=(color=red) name='Duration' nomissinggroup attrid=Duration;
highlow y=No_ low=start2 high=end2 / group=Duration lineattrs=(thickness=2 pattern=solid)
lineattrs=(color=blue) name='Duration' nomissinggroup attrid=Duration;
highlow y=No_ low=start3 high=end3 / group=Duration lineattrs=(thickness=2 pattern=solid)
lineattrs=(color=black) name='Duration' nomissinggroup attrid=Duration;
scatter X=start1 Y=No_ /markerattrs=(symbol=trianglefilled size=8 color=red) name='a' legendlabel='Mild Start';
scatter X=end1 Y=No_ /markerattrs=(symbol=circlefilled size=8 color=red) name='b' legendlabel='Mild End';
scatter X=start2 Y=No_ /markerattrs=(symbol=trianglefilled size=8 color=blue) name='c' legendlabel='Moderate Start';
scatter X=end2 Y=No_ /markerattrs=(symbol=circlefilled size=8 color=blue) name='d' legendlabel='Moderate End';
scatter X=start3 Y=No_ /markerattrs=(symbol=trianglefilled size=8 color=black) name='e' legendlabel='Severe Start';
scatter X=end3 Y=No_ /markerattrs=(symbol=circlefilled size=8 color=black) name='f' legendlabel='Severe End';
yaxis type=discrete display=(noticks) fitpolicy=none labelpos=top;
xaxis type=linear label="Week" values=(0 to 12 by 1);
keylegend 'drug' / title='Treatment Group';
keylegend 'a' 'b' 'c' 'd' 'e' 'f'/ noborder location=inside position=bottomright across=1;
run;
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Thank you, Sanjay. It is really helpful for me.
I set all start values as zero since this is my first time to generate Swimmer plot and also PROC SGPLOT.
In my point of view,
highlow y=No_ low=low high=line_end / group=drug attrid=A type=bar fill nooutline transparency=0.5
name='drug';this code is to determine the starting point and the end of swimmer lane. And the "scatter" code is to mark the points on the lane.
So, it will become complicated by setting different start values. Although, separating ids (113, 113A, 113B).. is the way to resolve the overlapping, I need to show one person by one swimmer lane even if there're multiple events of him/ her. Is it possible?
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Sure. Each lane is defined by a specific unique id. For ids with multiple events, create unique ids like 113a, 113b, 113c, to get different lanes. The label for each lane can be from another column, 'SubjId'. The displayed label can be 113 for each of the 3 lanes.
So, you need a unique column, say ID. Then you need another column say 'SubjId' which will contain the same value for all 3 values (113). Then display the SubjId using "LowLabel" option in the Highlow plot. Turn off the Y axis tick values.
highlow y=Id low=low high=line_end / group=drug attrid=A type=bar fill nooutline
transparency=0.5 name='drug' lowlabel=SubjId; yaxis display=(novalues nolabel);
- Tags:
- swimmer plot
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Hi Sanjay,
Sorry for the late reply. I got the heavy workload recently.
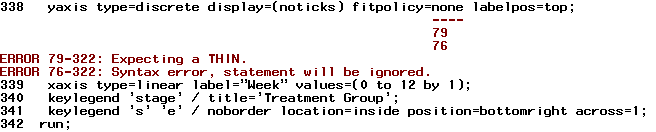
Well, I've just tried the code you provided, but it cannot work and popped-out the error message as follows. Could you please help to resolve, thank you.
When I'm using "fitpolicy=none labelpos=top" at the same time:
If I'm using only "labelpos=top" to show the label of y-axis:
Besides, can I ask about the function of "nocycleattrs". Thank you very much.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
So...is there any better way to show every tick mark value on the y-axis?
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
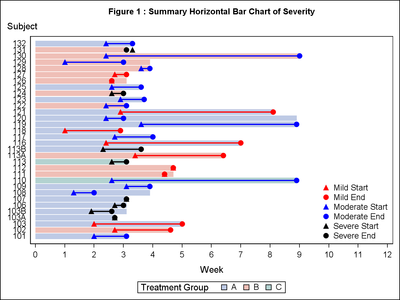
Hi John,
There are at least two solutions to help you to view each subject id.
One of them is to change the dimensions of the image, so that it is portrait instead of landscape. This will give you more space on the yaxis I can be done by simply changing the options within ods graphics (I modified Sanjay's earlier example):
ods graphics / reset imagename='Swimmer_portrait' height=6.67in width = 5in;
Another option, as Sanjay also said is to get rid of the labels on the y-axis and add the subjects labels at the end of the bar chart.
The following code, can help you achieve that: Basically the dataset has been updated to include the position where the subject labels should go, and then this label has been added to the end of the barchart using a new highlow statement - just because some of the start values, where after the end values.
* Make labels on bars;
data test2;
set test;
max_end = max(line_end, end1, end2, end3, start1, start2, start3);
label_x_axis = max_end + 0.1;
run;
ods listing gpath='C:\Users\yex7977\Communities\Image' image_dpi=200;
ods graphics / reset imagename='Swimmer_label';
title h=1 'Figure 1 : Summary Horizontal Bar Chart of Severity';
proc sgplot data=test2 nocycleattrs;
* label;
highlow y=No_ low=low high=label_x_axis / attrid=A type=bar lineattrs=(color=white) nofill outline transparency=0.5
name='drug' highlabel=No_;
highlow y=No_ low=low high=line_end / group=drug attrid=A type=bar fill nooutline transparency=0.5
name='drug';
highlow y=No_ low=start1 high=end1 / group=Duration lineattrs=(thickness=2 pattern=solid)
lineattrs=(color=red) name='Duration' nomissinggroup attrid=Duration;
highlow y=No_ low=start2 high=end2 / group=Duration lineattrs=(thickness=2 pattern=solid)
lineattrs=(color=blue) name='Duration' nomissinggroup attrid=Duration;
highlow y=No_ low=start3 high=end3 / group=Duration lineattrs=(thickness=2 pattern=solid)
lineattrs=(color=black) name='Duration' nomissinggroup attrid=Duration;
scatter X=start1 Y=No_ /markerattrs=(symbol=trianglefilled size=8 color=red) name='a' legendlabel='Mild Start';
scatter X=end1 Y=No_ /markerattrs=(symbol=circlefilled size=8 color=red) name='b' legendlabel='Mild End';
scatter X=start2 Y=No_ /markerattrs=(symbol=trianglefilled size=8 color=blue) name='c' legendlabel='Moderate Start';
scatter X=end2 Y=No_ /markerattrs=(symbol=circlefilled size=8 color=blue) name='d' legendlabel='Moderate End';
scatter X=start3 Y=No_ /markerattrs=(symbol=trianglefilled size=8 color=black) name='e' legendlabel='Severe Start';
scatter X=end3 Y=No_ /markerattrs=(symbol=circlefilled size=8 color=black) name='f' legendlabel='Severe End';
yaxis type=discrete display=NONE fitpolicy=none labelpos=top;
xaxis type=linear label="Week" values=(0 to 12 by 1);
keylegend 'drug' / title='Treatment Group';
keylegend 'a' 'b' 'c' 'd' 'e' 'f'/ noborder location=inside position=bottomright across=1;
run;

- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Thanks for your answer! The first one is the better for me. Maybe the graphs would look like landscape if I larger the width!
However, another issue is unsolved due to an older release of SAS as Sanjay said. So, the "label pos=" option cannot work.
Besides, I use SAS 9.3.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
If some options are not compiling for you, that means you have an older release of SAS.
NOCYCLEATTRS means the colors (and other attributes) will not continue to use increasing group indexes into the attribute list. For a graph that is built using multiple layers of "grouped" plots, it is better to explicitly control the colors using the Discrete Attributes Map.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
May I ask one more question? Is it possible to change the color of bars?
Learn how use the CAT functions in SAS to join values from multiple variables into a single value.
Find more tutorials on the SAS Users YouTube channel.
SAS Training: Just a Click Away
Ready to level-up your skills? Choose your own adventure.