- Home
- /
- Programming
- /
- Graphics
- /
- Re: How to change interval in x-axis for KM plot
- RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Hi there,
I have been struggling with this problem for a while.
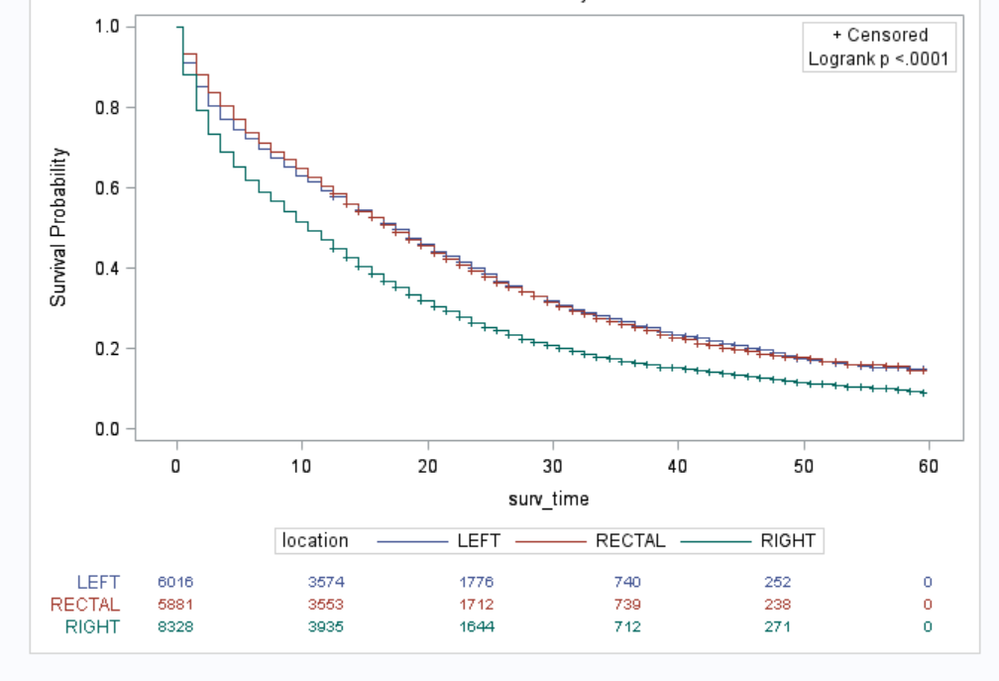
I run the PROC LIFETEST procedure below to get the KM plot. See the plot in the attachement. However, you will find that the time points along the x-axis are 0, 10, 20, 30, 40, 50, and 60. But I want the time interval to be 12, which can made the time points to be 0,12,24,36,48, and 60, so that the time points along x-axis is same as the time points for the number of at risk listing...I tried many options, even trying to revise PROC TEMPLATE. But none of them works......Does any one have resolved similar problem? Thanks in advance.
proc lifetest data=master1 intervals= 0 to 60 by 12
plots=survival(test atrisk(outside)=0 to 60 by 12);
by period;
strata location;
time surv_time*Censor(0);
run;
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
There are about 100 pages of documentation devoted to configuring the KM plot. Try modifying the example in the section "Modifying the axis" to use xOptions instead of yOptions .
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Hi Rick,
SAS gave me error message after I try this macro...see below
138 %ProvideSurvivalMacros
-
180
WARNING: Apparent invocation of macro PROVIDESURVIVALMACROS not resolved.
ERROR 180-322: Statement is not valid or it is used out of proper order.
139
140 %let xOptions = label="Survival Time"
141 linearopts=(viewmin=0 viewmax=60
142 tickvaluelist=(0 12 24 36 48 60));
143
144 %CompileSurvivalTemplates
WARNING: Apparent invocation of macro COMPILESURVIVALTEMPLATES not resolved.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Here's some old code I had- changed it to months. You can read through the comments and make changes as required.
/*SAS PROC LIFETEST Survival Plot Template
Modifed by: F. Khurshed
Date: July 27, 2011
Description: I modified the basic GTL plot to
1) Changed the thickness of the lines plotted
2) Changed the X axis markers to 12 month intervals
3) Changed the Y axis markers to 0.25 level increments
4) Changed the label of the legend to be a macro variable called legend_label. This needs to be specified prior to output
5) Changed the Y axis markers to be formatted to percent using the tickvalueformat option. (2012-05-01 FK)
*/
*Template appears in log;
proc template;
define statgraph Stat.Lifetest.Graphics.Productlimitsurvival;
dynamic NStrata xName plotAtRisk plotCensored plotCL plotHW plotEP labelCL labelHW labelEP maxTime method
StratumID classAtRisk plotBand plotTest GroupName yMin Transparency SecondTitle TestName pValue;
/*My Macro Variables*/
mvar legend_label;
BeginGraph;
if (NSTRATA=1)
if (EXISTS(STRATUMID))
entrytitle "Kaplan-Meier " "Survival Estimate" " for " STRATUMID;
else
entrytitle "Kaplan-Meier " "Survival Estimate";
endif;
if (PLOTATRISK)
entrytitle "with Number of Subjects at Risk" / textattrs=GRAPHVALUETEXT;
endif;
layout overlay / xaxisopts=(shortlabel=XNAME offsetmin=.05 linearopts=(viewmax=MAXTIME /*Modifying X axis values here, need to change below as well*/ tickvaluelist=(0 12 24 36 48 60 72 84 96 108 120))) yaxisopts=(label=
"Survival Probability" shortlabel="Survival" linearopts=(viewmin=0 viewmax=1 tickvaluelist=(0 .25 .5 .75
1.0) tickvalueformat=percent8.1 /*Modifying Y axis values here, need to change below as well*/));
/*Add a reference line for the media*/
*referenceline y=0.5/lineattrs=(color=red pattern=2);
if (PLOTHW=1 AND PLOTEP=0)
bandplot LimitUpper=HW_UCL LimitLower=HW_LCL x=TIME / modelname="Survival" fillattrs=GRAPHCONFIDENCE
name="HW" legendlabel=LABELHW;
endif;
if (PLOTHW=0 AND PLOTEP=1)
bandplot LimitUpper=EP_UCL LimitLower=EP_LCL x=TIME / modelname="Survival" fillattrs=GRAPHCONFIDENCE
name="EP" legendlabel=LABELEP;
endif;
if (PLOTHW=1 AND PLOTEP=1)
bandplot LimitUpper=HW_UCL LimitLower=HW_LCL x=TIME / modelname="Survival" fillattrs=GRAPHDATA1
datatransparency=.55 name="HW" legendlabel=LABELHW;
bandplot LimitUpper=EP_UCL LimitLower=EP_LCL x=TIME / modelname="Survival" fillattrs=GRAPHDATA2
datatransparency=.55 name="EP" legendlabel=LABELEP;
endif;
if (PLOTCL=1)
if (PLOTHW=1 OR PLOTEP=1)
bandplot LimitUpper=SDF_UCL LimitLower=SDF_LCL x=TIME / modelname="Survival" display=(outline)
outlineattrs=GRAPHPREDICTIONLIMITS name="CL" legendlabel=LABELCL;
else
bandplot LimitUpper=SDF_UCL LimitLower=SDF_LCL x=TIME / modelname="Survival" fillattrs=GRAPHCONFIDENCE
name="CL" legendlabel=LABELCL;
endif;
endif;
stepplot y=SURVIVAL x=TIME / name="Survival" rolename=(_tip1=ATRISK _tip2=EVENT) tip=(y x Time _tip1
_tip2) legendlabel="Survival" lineattrs=(thickness=2);
if (PLOTCENSORED=1)
scatterplot y=CENSORED x=TIME / markerattrs=(symbol=plus size=8px) name="Censored" legendlabel="Censored";
endif;
if (PLOTCL=1 OR PLOTHW=1 OR PLOTEP=1)
discretelegend "Censored" "CL" "HW" "EP" / location=outside halign=center;
else
if (PLOTCENSORED=1)
discretelegend "Censored" / location=inside autoalign=(topright bottomleft);
endif;
endif;
if (PLOTATRISK=1)
innermargin / align=bottom;
blockplot x=TATRISK block=ATRISK / repeatedvalues=true display=(values) valuehalign=start
valuefitpolicy=truncate labelposition=left labelattrs=GRAPHVALUETEXT valueattrs=GRAPHDATATEXT (size
=7pt) includemissingclass=false ;
endinnermargin;
endif;
endlayout;
else
entrytitle "Kaplan-Meier " "Survival Estimates";
layout overlay / xaxisopts=(shortlabel=XNAME offsetmin=.05 linearopts=(viewmax=MAXTIME tickvaluelist=(0 12 24 36 48 60 72 84 96 108 120) /*Modifying the X values on graph here, need to change above as well*/)) yaxisopts=(label=
"Survival Probability" shortlabel="Survival" linearopts=(viewmin=0 viewmax=1 tickvaluelist=(0 .25 .5 .75
1.0) tickvalueformat=percent8.1 /*Modifying the Y Axis values here, need to change above as well*/));
/*Add a reference line for the media*/
*referenceline y=0.5/lineattrs=(color=red pattern=2);
if (PLOTHW)
bandplot LimitUpper=HW_UCL LimitLower=HW_LCL x=TIME / group=STRATUM index=STRATUMNUM modelname=
"Survival" datatransparency=Transparency;
endif;
if (PLOTEP)
bandplot LimitUpper=EP_UCL LimitLower=EP_LCL x=TIME / group=STRATUM index=STRATUMNUM modelname=
"Survival" datatransparency=Transparency;
endif;
if (PLOTCL)
if (PLOTBAND)
bandplot LimitUpper=SDF_UCL LimitLower=SDF_LCL x=TIME / group=STRATUM index=STRATUMNUM modelname=
"Survival" display=(outline);
else
bandplot LimitUpper=SDF_UCL LimitLower=SDF_LCL x=TIME / group=STRATUM index=STRATUMNUM modelname=
"Survival" datatransparency=Transparency;
endif;
endif;
stepplot y=SURVIVAL x=TIME / group=STRATUM index=STRATUMNUM name="Survival" rolename=(_tip1=ATRISK _tip2=
EVENT) tip=(y x Time _tip1 _tip2) lineattrs=(thickness=2);
if (PLOTCENSORED)
scatterplot y=CENSORED x=TIME / group=STRATUM index=STRATUMNUM markerattrs=(symbol=plus size=8px);
endif;
if (PLOTATRISK)
innermargin / align=bottom;
blockplot x=TATRISK block=ATRISK / class=STRATUM repeatedvalues=true display=(label values)
valuehalign=start valuefitpolicy=truncate labelposition=left labelattrs=STRATUMID valueattrs=
GRAPHDATATEXT (size=7pt) includemissingclass=false;
endinnermargin;
endif;
DiscreteLegend "Survival" / title=legend_label location=outside;
if (PLOTCENSORED)
if (PLOTTEST)
layout gridded / rows=2 autoalign=(TOPRIGHT BOTTOMLEFT TOP BOTTOM) border=false BackgroundColor=
GraphWalls:Color Opaque=true;
entry "+ Censored";
if (PVALUE < .0001)
entry TESTNAME " p " eval (PUT(PVALUE, PVALUE6.4));
else
entry TESTNAME " p=" eval (PUT(PVALUE, PVALUE6.4));
endif;
endlayout;
else
layout gridded / rows=1 autoalign=(TOPRIGHT BOTTOMLEFT TOP BOTTOM) border=false BackgroundColor=
GraphWalls:Color Opaque=true;
entry "+ Censored";
endlayout;
endif;
else
if (PLOTTEST)
layout gridded / rows=1 autoalign=(TOPRIGHT BOTTOMLEFT TOP BOTTOM) border=false BackgroundColor=
GraphWalls:Color Opaque=true;
if (PVALUE < .0001)
entry TESTNAME " p " eval (PUT(PVALUE, PVALUE6.4));
else
entry TESTNAME " p=" eval (PUT(PVALUE, PVALUE6.4));
endif;
endlayout;
endif;
endif;
endlayout;
endif;
EndGraph;
end;
run;
https://gist.github.com/statgeek/4f8ccc4ee603b41371e5
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Hi Reeza,
Thanks for sharing. However, this still does not work...It is very very weird that PROC TEMPLATE does not work the plot on my end...
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
What version of SAS are you running?
Post your code and log.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Hi Reeza,
I am using SAS 9.3. See the code in log. Thanks!
101 proc template;
102 define statgraph Stat.Lifetest.Graphics.Productlimitsurvival;
103 dynamic NStrata xName plotAtRisk plotCensored plotCL plotHW plotEP labelCL labelHW labelEP
103! maxTime method
104 StratumID classAtRisk plotBand plotTest GroupName yMin Transparency SecondTitle
104! TestName pValue;
105
106 /*My Macro Variables*/
107 mvar legend_label;
108 BeginGraph;
109 if (NSTRATA=1)
110 if (EXISTS(STRATUMID))
111 entrytitle "Kaplan-Meier " "Survival Estimate" " for " STRATUMID;
112 else
113 entrytitle "Kaplan-Meier " "Survival Estimate";
114 endif;
115 if (PLOTATRISK)
116 entrytitle "with Number of Subjects at Risk" / textattrs=GRAPHVALUETEXT;
117 endif;
118 layout overlay / xaxisopts=(shortlabel=XNAME offsetmin=.05 linearopts=(viewmax=MAXTIME
118! /*Modifying X axis values here, need to change below as well*/ tickvaluelist=(0 12 24 36 48
118! 60 72 84 96 108 120))) yaxisopts=(label=
119 "Survival Probability" shortlabel="Survival" linearopts=(viewmin=0 viewmax=1
119! tickvaluelist=(0 .25 .5 .75
120 1.0) tickvalueformat=percent8.1 /*Modifying Y axis values here, need to change below
120! as well*/));
121
122 /*Add a reference line for the media*/
123 *referenceline y=0.5/lineattrs=(color=red pattern=2);
124 if (PLOTHW=1 AND PLOTEP=0)
125 bandplot LimitUpper=HW_UCL LimitLower=HW_LCL x=TIME / modelname="Survival"
125! fillattrs=GRAPHCONFIDENCE
126 name="HW" legendlabel=LABELHW;
127 endif;
128 if (PLOTHW=0 AND PLOTEP=1)
129 bandplot LimitUpper=EP_UCL LimitLower=EP_LCL x=TIME / modelname="Survival"
129! fillattrs=GRAPHCONFIDENCE
130 name="EP" legendlabel=LABELEP;
131 endif;
132 if (PLOTHW=1 AND PLOTEP=1)
133 bandplot LimitUpper=HW_UCL LimitLower=HW_LCL x=TIME / modelname="Survival"
133! fillattrs=GRAPHDATA1
134 datatransparency=.55 name="HW" legendlabel=LABELHW;
135 bandplot LimitUpper=EP_UCL LimitLower=EP_LCL x=TIME / modelname="Survival"
135! fillattrs=GRAPHDATA2
136 datatransparency=.55 name="EP" legendlabel=LABELEP;
137 endif;
138 if (PLOTCL=1)
139 if (PLOTHW=1 OR PLOTEP=1)
140 bandplot LimitUpper=SDF_UCL LimitLower=SDF_LCL x=TIME / modelname="Survival"
140! display=(outline)
141 outlineattrs=GRAPHPREDICTIONLIMITS name="CL" legendlabel=LABELCL;
142 else
143 bandplot LimitUpper=SDF_UCL LimitLower=SDF_LCL x=TIME / modelname="Survival"
143! fillattrs=GRAPHCONFIDENCE
144 name="CL" legendlabel=LABELCL;
145 endif;
146 endif;
147 stepplot y=SURVIVAL x=TIME / name="Survival" rolename=(_tip1=ATRISK _tip2=EVENT)
147! tip=(y x Time _tip1
148 _tip2) legendlabel="Survival" lineattrs=(thickness=2);
149 if (PLOTCENSORED=1)
150 scatterplot y=CENSORED x=TIME / markerattrs=(symbol=plus size=8px)
150! name="Censored" legendlabel="Censored";
151 endif;
152 if (PLOTCL=1 OR PLOTHW=1 OR PLOTEP=1)
153 discretelegend "Censored" "CL" "HW" "EP" / location=outside halign=center;
154 else
155 if (PLOTCENSORED=1)
156 discretelegend "Censored" / location=inside autoalign=(topright bottomleft);
157 endif;
158 endif;
159 if (PLOTATRISK=1)
160 innermargin / align=bottom;
161 blockplot x=TATRISK block=ATRISK / repeatedvalues=true display=(values)
161! valuehalign=start
162 valuefitpolicy=truncate labelposition=left labelattrs=GRAPHVALUETEXT
162! valueattrs=GRAPHDATATEXT (size
163 =7pt) includemissingclass=false ;
164 endinnermargin;
165 endif;
166 endlayout;
167 else
168 entrytitle "Kaplan-Meier " "Survival Estimates";
169 layout overlay / xaxisopts=(shortlabel=XNAME offsetmin=.05 linearopts=(viewmax=MAXTIME
169! tickvaluelist=(0 12 24 36 48 60 72 84 96 108 120) /*Modifying the X values on graph here,
169! need to change above as well*/)) yaxisopts=(label=
170 "Survival Probability" shortlabel="Survival" linearopts=(viewmin=0 viewmax=1
170! tickvaluelist=(0 .25 .5 .75
171 1.0) tickvalueformat=percent8.1 /*Modifying the Y Axis values here, need to change
171! above as well*/));
172
173 /*Add a reference line for the media*/
174 *referenceline y=0.5/lineattrs=(color=red pattern=2);
175 if (PLOTHW)
176 bandplot LimitUpper=HW_UCL LimitLower=HW_LCL x=TIME / group=STRATUM
176! index=STRATUMNUM modelname=
177 "Survival" datatransparency=Transparency;
178 endif;
179 if (PLOTEP)
180 bandplot LimitUpper=EP_UCL LimitLower=EP_LCL x=TIME / group=STRATUM
180! index=STRATUMNUM modelname=
181 "Survival" datatransparency=Transparency;
182 endif;
183 if (PLOTCL)
184 if (PLOTBAND)
185 bandplot LimitUpper=SDF_UCL LimitLower=SDF_LCL x=TIME / group=STRATUM
185! index=STRATUMNUM modelname=
186 "Survival" display=(outline);
187 else
188 bandplot LimitUpper=SDF_UCL LimitLower=SDF_LCL x=TIME / group=STRATUM
188! index=STRATUMNUM modelname=
189 "Survival" datatransparency=Transparency;
190 endif;
191 endif;
192 stepplot y=SURVIVAL x=TIME / group=STRATUM index=STRATUMNUM name="Survival"
192! rolename=(_tip1=ATRISK _tip2=
193 EVENT) tip=(y x Time _tip1 _tip2) lineattrs=(thickness=2);
194 if (PLOTCENSORED)
195 scatterplot y=CENSORED x=TIME / group=STRATUM index=STRATUMNUM
195! markerattrs=(symbol=plus size=8px);
196 endif;
197 if (PLOTATRISK)
198 innermargin / align=bottom;
199 blockplot x=TATRISK block=ATRISK / class=STRATUM repeatedvalues=true
199! display=(label values)
200 valuehalign=start valuefitpolicy=truncate labelposition=left
200! labelattrs=STRATUMID valueattrs=
201 GRAPHDATATEXT (size=7pt) includemissingclass=false;
202 endinnermargin;
203 endif;
204 DiscreteLegend "Survival" / title=legend_label location=outside;
205 if (PLOTCENSORED)
206 if (PLOTTEST)
207 layout gridded / rows=2 autoalign=(TOPRIGHT BOTTOMLEFT TOP BOTTOM) border=false
207! BackgroundColor=
208 GraphWalls:Color Opaque=true;
209 entry "+ Censored";
210 if (PVALUE < .0001)
211 entry TESTNAME " p " eval (PUT(PVALUE, PVALUE6.4));
212 else
213 entry TESTNAME " p=" eval (PUT(PVALUE, PVALUE6.4));
214 endif;
215 endlayout;
216 else
217 layout gridded / rows=1 autoalign=(TOPRIGHT BOTTOMLEFT TOP BOTTOM) border=false
217! BackgroundColor=
218 GraphWalls:Color Opaque=true;
219 entry "+ Censored";
220 endlayout;
221 endif;
222 else
223 if (PLOTTEST)
224 layout gridded / rows=1 autoalign=(TOPRIGHT BOTTOMLEFT TOP BOTTOM) border=false
224! BackgroundColor=
225 GraphWalls:Color Opaque=true;
226 if (PVALUE < .0001)
227 entry TESTNAME " p " eval (PUT(PVALUE, PVALUE6.4));
228 else
229 entry TESTNAME " p=" eval (PUT(PVALUE, PVALUE6.4));
230 endif;
231 endlayout;
232 endif;
233 endif;
234 endlayout;
235 endif;
236 EndGraph;
237 end;
NOTE: Overwriting existing template/link: Stat.Lifetest.Graphics.Productlimitsurvival
NOTE: STATGRAPH 'Stat.Lifetest.Graphics.Productlimitsurvival' has been saved to: SASUSER.TEMPLAT
238 run;
NOTE: PROCEDURE TEMPLATE used (Total process time):
real time 0.09 seconds
cpu time 0.09 seconds
239 proc lifetest data=master1 intervals= 0 to 60 by 12
240 plots=survival(test atrisk(outside)=0 to 60 by 12);
241 by period;
242 strata location;
243 time surv_time*Censor(0);
244 run;
NOTE: The LOGLOG transform is used to compute the confidence limits for the quartiles of the
survivor distribution. To suppress using this transform, specify CONFTYPE=LINEAR in the
PROC LIFETEST statement.
NOTE: PROCEDURE LIFETEST used (Total process time):
real time 1.85 seconds
cpu time 0.85 seconds
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Verify which template your graph is using - remove the atrisk and test statements, leaving survival alone and see if that works. Based on your specifications, it may be using a different template.
Use
ODS TRACE ON;
code
ODS TRACE OFF;
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Hi Reeza,
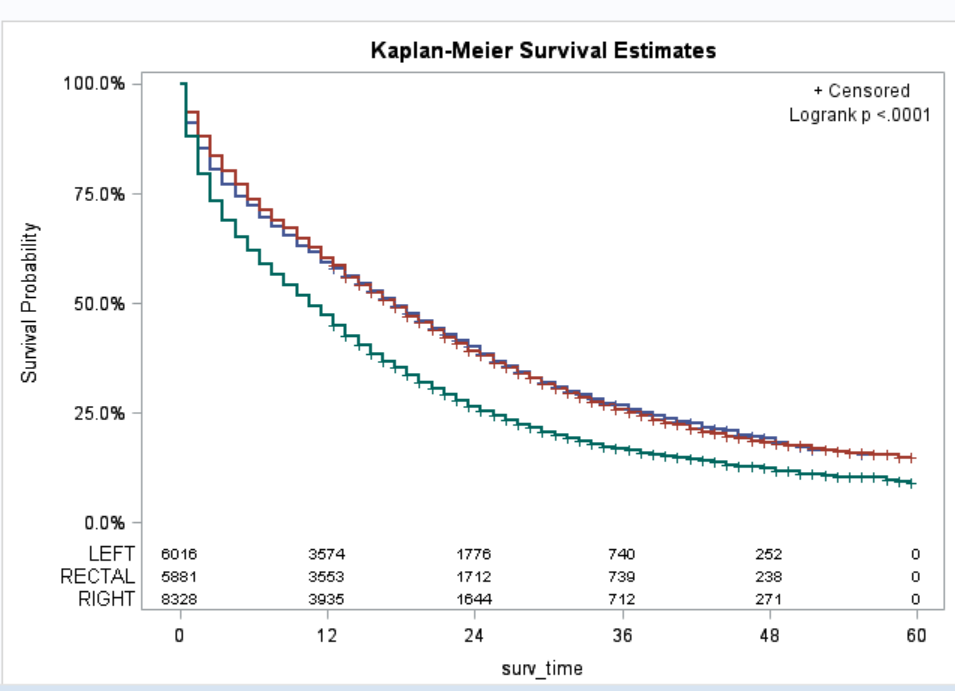
You are right, my template name is Stat.Lifetest.Graphics.Productlimitsurvival2. After I updated the name, the code works. But now, the list of number at risk is within the plot..See the attachement. If I want it to be outside of the plot, like below the x-axis, could you mind telling me from which line of your code that I can start to revise. Thank you very much!
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
@ @ Tiny_Kane Is there a way to modify this macro for doing CIF plot using proc phreg?
@Tiny_Kane wrote:Hi Reeza,
I am using SAS 9.3. See the code in log. Thanks!
101 proc template;
102 define statgraph Stat.Lifetest.Graphics.Productlimitsurvival;
103 dynamic NStrata xName plotAtRisk plotCensored plotCL plotHW plotEP labelCL labelHW labelEP
103! maxTime method
104 StratumID classAtRisk plotBand plotTest GroupName yMin Transparency SecondTitle
104! TestName pValue;
105
106 /*My Macro Variables*/
107 mvar legend_label;
108 BeginGraph;
109 if (NSTRATA=1)
110 if (EXISTS(STRATUMID))
111 entrytitle "Kaplan-Meier " "Survival Estimate" " for " STRATUMID;
112 else
113 entrytitle "Kaplan-Meier " "Survival Estimate";
114 endif;
115 if (PLOTATRISK)
116 entrytitle "with Number of Subjects at Risk" / textattrs=GRAPHVALUETEXT;
117 endif;
118 layout overlay / xaxisopts=(shortlabel=XNAME offsetmin=.05 linearopts=(viewmax=MAXTIME
118! /*Modifying X axis values here, need to change below as well*/ tickvaluelist=(0 12 24 36 48
118! 60 72 84 96 108 120))) yaxisopts=(label=
119 "Survival Probability" shortlabel="Survival" linearopts=(viewmin=0 viewmax=1
119! tickvaluelist=(0 .25 .5 .75
120 1.0) tickvalueformat=percent8.1 /*Modifying Y axis values here, need to change below
120! as well*/));
121
122 /*Add a reference line for the media*/
123 *referenceline y=0.5/lineattrs=(color=red pattern=2);
124 if (PLOTHW=1 AND PLOTEP=0)
125 bandplot LimitUpper=HW_UCL LimitLower=HW_LCL x=TIME / modelname="Survival"
125! fillattrs=GRAPHCONFIDENCE
126 name="HW" legendlabel=LABELHW;
127 endif;
128 if (PLOTHW=0 AND PLOTEP=1)
129 bandplot LimitUpper=EP_UCL LimitLower=EP_LCL x=TIME / modelname="Survival"
129! fillattrs=GRAPHCONFIDENCE
130 name="EP" legendlabel=LABELEP;
131 endif;
132 if (PLOTHW=1 AND PLOTEP=1)
133 bandplot LimitUpper=HW_UCL LimitLower=HW_LCL x=TIME / modelname="Survival"
133! fillattrs=GRAPHDATA1
134 datatransparency=.55 name="HW" legendlabel=LABELHW;
135 bandplot LimitUpper=EP_UCL LimitLower=EP_LCL x=TIME / modelname="Survival"
135! fillattrs=GRAPHDATA2
136 datatransparency=.55 name="EP" legendlabel=LABELEP;
137 endif;
138 if (PLOTCL=1)
139 if (PLOTHW=1 OR PLOTEP=1)
140 bandplot LimitUpper=SDF_UCL LimitLower=SDF_LCL x=TIME / modelname="Survival"
140! display=(outline)
141 outlineattrs=GRAPHPREDICTIONLIMITS name="CL" legendlabel=LABELCL;
142 else
143 bandplot LimitUpper=SDF_UCL LimitLower=SDF_LCL x=TIME / modelname="Survival"
143! fillattrs=GRAPHCONFIDENCE
144 name="CL" legendlabel=LABELCL;
145 endif;
146 endif;
147 stepplot y=SURVIVAL x=TIME / name="Survival" rolename=(_tip1=ATRISK _tip2=EVENT)
147! tip=(y x Time _tip1
148 _tip2) legendlabel="Survival" lineattrs=(thickness=2);
149 if (PLOTCENSORED=1)
150 scatterplot y=CENSORED x=TIME / markerattrs=(symbol=plus size=8px)
150! name="Censored" legendlabel="Censored";
151 endif;
152 if (PLOTCL=1 OR PLOTHW=1 OR PLOTEP=1)
153 discretelegend "Censored" "CL" "HW" "EP" / location=outside halign=center;
154 else
155 if (PLOTCENSORED=1)
156 discretelegend "Censored" / location=inside autoalign=(topright bottomleft);
157 endif;
158 endif;
159 if (PLOTATRISK=1)
160 innermargin / align=bottom;
161 blockplot x=TATRISK block=ATRISK / repeatedvalues=true display=(values)
161! valuehalign=start
162 valuefitpolicy=truncate labelposition=left labelattrs=GRAPHVALUETEXT
162! valueattrs=GRAPHDATATEXT (size
163 =7pt) includemissingclass=false ;
164 endinnermargin;
165 endif;
166 endlayout;
167 else
168 entrytitle "Kaplan-Meier " "Survival Estimates";
169 layout overlay / xaxisopts=(shortlabel=XNAME offsetmin=.05 linearopts=(viewmax=MAXTIME
169! tickvaluelist=(0 12 24 36 48 60 72 84 96 108 120) /*Modifying the X values on graph here,
169! need to change above as well*/)) yaxisopts=(label=
170 "Survival Probability" shortlabel="Survival" linearopts=(viewmin=0 viewmax=1
170! tickvaluelist=(0 .25 .5 .75
171 1.0) tickvalueformat=percent8.1 /*Modifying the Y Axis values here, need to change
171! above as well*/));
172
173 /*Add a reference line for the media*/
174 *referenceline y=0.5/lineattrs=(color=red pattern=2);
175 if (PLOTHW)
176 bandplot LimitUpper=HW_UCL LimitLower=HW_LCL x=TIME / group=STRATUM
176! index=STRATUMNUM modelname=
177 "Survival" datatransparency=Transparency;
178 endif;
179 if (PLOTEP)
180 bandplot LimitUpper=EP_UCL LimitLower=EP_LCL x=TIME / group=STRATUM
180! index=STRATUMNUM modelname=
181 "Survival" datatransparency=Transparency;
182 endif;
183 if (PLOTCL)
184 if (PLOTBAND)
185 bandplot LimitUpper=SDF_UCL LimitLower=SDF_LCL x=TIME / group=STRATUM
185! index=STRATUMNUM modelname=
186 "Survival" display=(outline);
187 else
188 bandplot LimitUpper=SDF_UCL LimitLower=SDF_LCL x=TIME / group=STRATUM
188! index=STRATUMNUM modelname=
189 "Survival" datatransparency=Transparency;
190 endif;
191 endif;
192 stepplot y=SURVIVAL x=TIME / group=STRATUM index=STRATUMNUM name="Survival"
192! rolename=(_tip1=ATRISK _tip2=
193 EVENT) tip=(y x Time _tip1 _tip2) lineattrs=(thickness=2);
194 if (PLOTCENSORED)
195 scatterplot y=CENSORED x=TIME / group=STRATUM index=STRATUMNUM
195! markerattrs=(symbol=plus size=8px);
196 endif;
197 if (PLOTATRISK)
198 innermargin / align=bottom;
199 blockplot x=TATRISK block=ATRISK / class=STRATUM repeatedvalues=true
199! display=(label values)
200 valuehalign=start valuefitpolicy=truncate labelposition=left
200! labelattrs=STRATUMID valueattrs=
201 GRAPHDATATEXT (size=7pt) includemissingclass=false;
202 endinnermargin;
203 endif;
204 DiscreteLegend "Survival" / title=legend_label location=outside;
205 if (PLOTCENSORED)
206 if (PLOTTEST)
207 layout gridded / rows=2 autoalign=(TOPRIGHT BOTTOMLEFT TOP BOTTOM) border=false
207! BackgroundColor=
208 GraphWalls:Color Opaque=true;
209 entry "+ Censored";
210 if (PVALUE < .0001)
211 entry TESTNAME " p " eval (PUT(PVALUE, PVALUE6.4));
212 else
213 entry TESTNAME " p=" eval (PUT(PVALUE, PVALUE6.4));
214 endif;
215 endlayout;
216 else
217 layout gridded / rows=1 autoalign=(TOPRIGHT BOTTOMLEFT TOP BOTTOM) border=false
217! BackgroundColor=
218 GraphWalls:Color Opaque=true;
219 entry "+ Censored";
220 endlayout;
221 endif;
222 else
223 if (PLOTTEST)
224 layout gridded / rows=1 autoalign=(TOPRIGHT BOTTOMLEFT TOP BOTTOM) border=false
224! BackgroundColor=
225 GraphWalls:Color Opaque=true;
226 if (PVALUE < .0001)
227 entry TESTNAME " p " eval (PUT(PVALUE, PVALUE6.4));
228 else
229 entry TESTNAME " p=" eval (PUT(PVALUE, PVALUE6.4));
230 endif;
231 endlayout;
232 endif;
233 endif;
234 endlayout;
235 endif;
236 EndGraph;
237 end;
NOTE: Overwriting existing template/link: Stat.Lifetest.Graphics.Productlimitsurvival
NOTE: STATGRAPH 'Stat.Lifetest.Graphics.Productlimitsurvival' has been saved to: SASUSER.TEMPLAT
238 run;
NOTE: PROCEDURE TEMPLATE used (Total process time):
real time 0.09 seconds
cpu time 0.09 seconds
239 proc lifetest data=master1 intervals= 0 to 60 by 12
240 plots=survival(test atrisk(outside)=0 to 60 by 12);
241 by period;
242 strata location;
243 time surv_time*Censor(0);
244 run;
NOTE: The LOGLOG transform is used to compute the confidence limits for the quartiles of the
survivor distribution. To suppress using this transform, specify CONFTYPE=LINEAR in the
PROC LIFETEST statement.
NOTE: PROCEDURE LIFETEST used (Total process time):
real time 1.85 seconds
cpu time 0.85 seconds
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
@ Reeza Is there a way to modify this macro for doing CIF plot using proc phreg or proc lifetest? I used this code successfully to reformat the X axis for my survival curve but am now trying to change it for CIF
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Yes. You need to read the documentation, which includes instructions on how to download and use the macros.
Learn how use the CAT functions in SAS to join values from multiple variables into a single value.
Find more tutorials on the SAS Users YouTube channel.
SAS Training: Just a Click Away
Ready to level-up your skills? Choose your own adventure.



