- Home
- /
- Analytics
- /
- Stat Procs
- /
- mixed model specification
- RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
I have a dataset (see attached), which basically involves 4 treatments for a chemotherapy drug. Samples were taken from 2 biopsy locations, and biopsy were taken at 2 time points. So biopsy location and time are nested within subject, and thus each subject has 4 data points (from 2 biopsy locations and 2 time points). The objective is to study treatment difference.
time is 2-level factor (Day 4, Day 10)
biopsysite is 2-level factor (S1 and S2)
trt is 4-level factor (A, B, C, D)
An exmple part of the dataet looks like:
pid trt time biopsysite y age
P2 A DAY 4 S1 -0.122 49
P2 A DAY 4 S2 -0.442 49
P2 A DAY 10 S1 0.229 49
P2 A DAY 10 S2 -0.007 49
P8 B DAY 4 S2 0.748 62
P8 B DAY 4 S1 0.086 62
P8 B DAY 10 S2 0.090 62
P8 B DAY 10 S1 0.076 62
:
:
:
I would like to fit a mixed model with age, time, trt and time by trt as fixed factor. "time" as repeated measure using AR1(), and "biopsysite" as random with compound symmetry covariance structure between the 2 biopsy locations.
How should I specify the mixed model? Is the following correct?
proc mixed;
class pid trt time biopsysite;
model y = age trt time trt*time / s;
random biopsysite / type=cs subject=pid g;
repeated / type=ar(1) subject=pid*biopsysite r;
lsmeans trt*time/cl pdiff;
run;
Or what about this one, does it make sense?
proc mixed data=test;
class time trt pid biopsysite;
model y=age time trt time*trt/s;
random biopsysite/type=cs g;
repeated pid / type=ar(1) r;
lsmeans trt*time / cl pdiff alpha=0.2;
run;
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
i am thinking
proc mixed data=test;
class time trt pid biopsysite;
model y=age time trt time*trt/s;
random biopsysite / type=cs subject=pid g;
repeated time / type=ar(1) subject=pid r;
lsmeans trt*time / cl pdiff alpha=0.2;
run;
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Thanks for suggestion. The reason I was using "subject=pid*biopsysite" in repeated statement was because the 2 time points thar were repeated were per each biopsy site, by specifying pid*biopsite intereaction, I was making sure the ar(1) covariance structure is applied to the 2 time point within biopsy site within subject. Accoording to MIXED manual:
"use the SUBJECT= option to define the blocks of 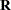 and the TYPE= option to define their covariance structure"
and the TYPE= option to define their covariance structure"
The block of R in my example is within biopsy site within subject.
Am I understanding this correctly?
Thanks
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
ah in that case do repeated time / type=ar(1) subject=biopsysite(pid)
the subject=biopsysite(pid) specifies that each biopsysite is nested with each pid. you may also want to try random intercept / type=un subject=pid that way you are fitting a random intercept for each person as well as an ar(1) covariance structure for the time points
this ultimately gives you a hybrid covariance structure. i can't think of the complete details off the top of my head (i'll have to reference back to my old school notes if you want more info)
cheers
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Thank you very much!
ANOVA, or Analysis Of Variance, is used to compare the averages or means of two or more populations to better understand how they differ. Watch this tutorial for more.
Find more tutorials on the SAS Users YouTube channel.


