- Home
- /
- Analytics
- /
- Stat Procs
- /
- Re: Help with PROC LIFETEST multiple comparison test results
- RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
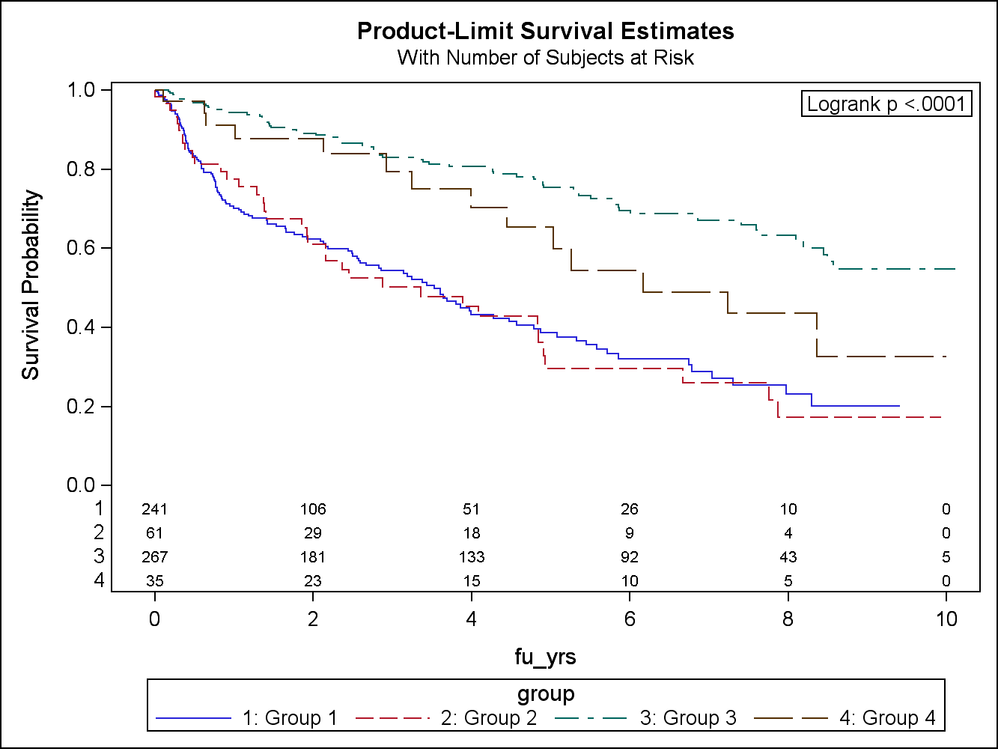
Can someone help me understand why my PROC LIFETEST results for multiple comparison tests are all significant given the Kaplan-Meier curves (in attached .png file)? Seems like groups 1 and 2 should NOT be significant. However, the paired comparison test shows up as significant (in attached .pdf). Here is the code I'm running:
proc lifetest data=getdata2 plots=s(atrisk=0 to 10 by 2 nocensor test) notable;
time fu_yrs*event(0);
strata group / diff=all adjust=bon;
run;
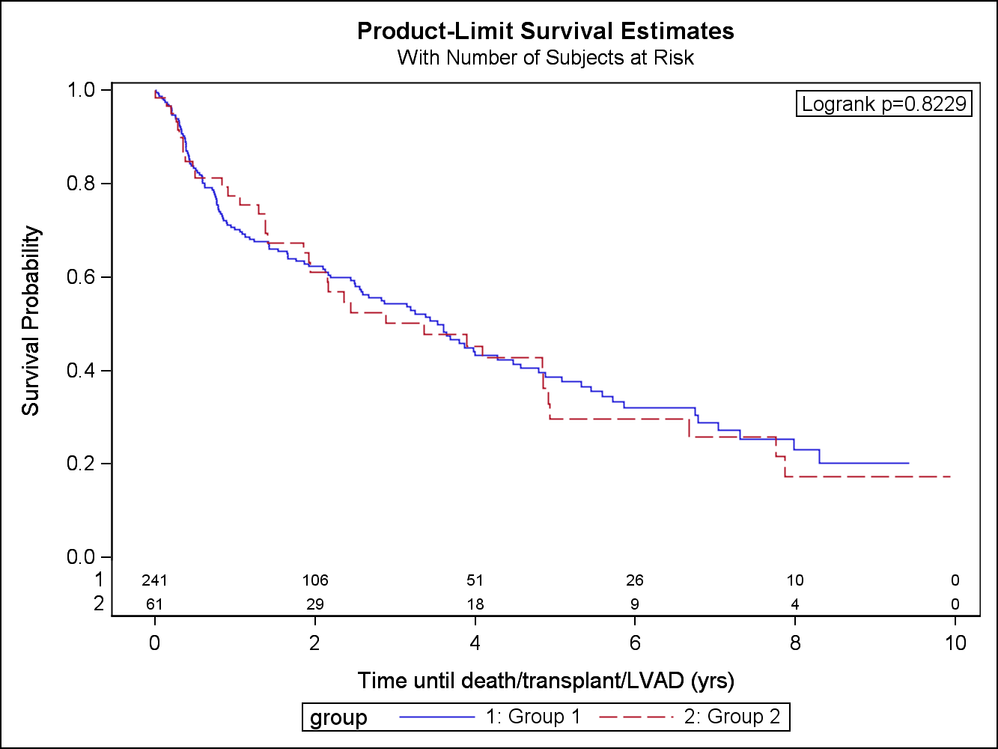
When I run a separate procedure and restrict the data to only groups 1 and 2, the log-rank test is highly non-significant (log-rank p=0.82) and what I would expect given the curves.
Please help me understand. Thanks!
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
log-rank test has most power when these two survival curve are parallel .
but From your Photo, It seems that you should use some non-parameter method Such as Wilcoxon Test to instead of Log-Rank test.
Ksharp
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Thanks Ksharp, I appreciate the response. The results of the Wilcoxon tests mirror that of the log-rank tests in that each paired comparison is significant. Yet the curves for groups 1 and 2 appear so similar and cross at various time points (early, mid, and late) that it just does not seem that these should be found to be statistically different. I'm still confused and am wondering if I should trust the DIFF=ALL option in the STRATA statement for PROC LIFETEST.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Sorry. Actually I am not very familiar with Survival Analysis.
There must be some specialist can reponse your problem. Like user : Reeza.
Ksharp
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
There's definitely wrong with something somewhere...
You have a lot of censoring in your data but I doubt that's the issue.
If you run the proc with only the 2 groups (ie group 1 and group 2) the raw p-value should equal the raw p-value in your adjustment for multiple comparison table, which it clearly doesn't according to what you're seeing.
Your data show's that two values are getting excluded due to missing observations, so you would need to make sure that the 2 are also being excluded for both analysis sets. I don't see the two observations being that influential regardless, but worth checking.
Can you post the plot that you get from running the two groups alone and your log from both of the procs?
Your best (and fastest) bet may be contact tech support though.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Hi Reeza,
Thanks for taking the time to help. Attached are the survival curves for just groups 1 and 2. Also attached is the log file from running the 4 group analysis first, then the 2 group analysis. I also included the output. I will follow up on those 2 missing observations and see if I can get those corrected.
If you don't see anything else, I probably will contact tech support. It does provide some comfort to me that I'm not the only one who thinks something is off. I'm not 100% convinced that it isn't me, though. ![]()
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Ok...I'd suggest contacting tech support.
This is the closest Note I could find to your issue which doesn't seem very relevant.
http://support.sas.com/kb/37/728.html
So you could try the sort and see if it's a similar issue depending on what version of SAS you're on.
Post the answer back to the forum please ![]()
Just a note you can use the timelist=0 to 10 by 2 to reduce your output some more so you don't have the long outputs of the survival tables as well.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Thanks Reeza for the timelist option suggestion. I will definitely use it in the future.
SAS support is really terrific about responding. They always seem to have a quick turn-around time and are extremely helpful. Below is the un-editted response I rec'd. It may well be as they suggest, but I still remain highly skeptical.
"Statistically significance has much to do with the sample size. If you have a large enough sample size, you can detect very small differences. Although the curves for Group1 and Group2 appears to be close to each other, the large sample sizes may still detect a difference. If you use the data for Group1 and Group2 alone, the insignificant difference could be due to a smaller sample size.
If you look at the chi-square values in the multiple comparison results, you see Group 3 is different from all other groups. And Group1 and Group2 pair and Group2 and Group4 pairs are less different. "
+ - - - - - -
The gist of the idea here is that your results are theoretically and practically possible to encounter and we don't suspect anything is amiss here. We have also been notified of situations where the overall test was significant but none of the pair-wise tests were significant - and this seemingly anomalous situation is also theoretically and practically possible as well.
Although not in the survival analysis realm, Milliken & Johnson(1992) in their "Analysis of Messy Data" book present a seemingly anomalous multiple comparisons example whereby means that are far apart were not significantly different but means that were closer together were significantly different. In their discussion, as I recall, the culprit was the varying/unbalanced sample sizes among the groups. I don't have access to Milliken & Johnson at present but I believe this example was in Chapter 3 at or near the end.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
I did notice that your group sizes are very different but I thought the following would hold...
If you run the proc with only the 2 groups (ie group 1 and group 2) the raw p-value should equal the raw p-value in your adjustment for multiple comparison table.
Message was edited by: Reeza It has to do with the variance estimates which change when you run a model with only two groups versus with all four groups. Compare your variance estimates for Group 1 and 2 from the model with all 4 groups (-6.5257) to the model with only two groups (-27.1545).
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Hi, I had the same problem. Does anyone have solutions for this problem? Should I trust p-value from 2 levels subset comparison or multiple comparison? thanks.
April 27 – 30 | Gaylord Texan | Grapevine, Texas
Registration is open
Walk in ready to learn. Walk out ready to deliver. This is the data and AI conference you can't afford to miss.
Register now and save with the early bird rate—just $795!
ANOVA, or Analysis Of Variance, is used to compare the averages or means of two or more populations to better understand how they differ. Watch this tutorial for more.
Find more tutorials on the SAS Users YouTube channel.


