- Home
- /
- Analytics
- /
- Stat Procs
- /
- Applying linear mixed model where the dependent variable is skewed
- RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Hi SAS Community,
I have a repeated measures study design where the dependent variable is calorie and the independent variables are day and disease severity. An example of the person-time dataset is shown below:
Subject Calorie Day Disease_severity
1 1000 1 Mild
1 1200 2 Mild
1 900 3 Mild
2 1800 1 Moderate
2 1600 2 Moderate
2 1700 3 Moderate
Since measures from the same subject are auto-correlated, I want to apply linear mixed model with day and disease severity as the independent variable, calories as the dependent variable and specifying a repeated statement for days.
proc mixed data=have;
title 'mixed model';
class subject day disease_severity;
model calorie=day disease_severity;
repeated day/subject=subject type=un;
lsmeans day;
run;
The issue is that the dependent variable is nonnormal and the measures of central tendency (mean vs median) are very different. Given the distribution, is it worth considering generalized linear mixed model. My understanding is that in generalized linear models the dependent variables can result in residuals that are normally distributed. I have three questions:
1. How can I evaluate if the dependent variable residual is nonnormal?
2. If generalized linear mixed model is appropriate, what distribution does the errors follow in this case? Is the error in this case part of the exponential family?
3. Should I consider other approaches such as quintile regression?
As always, your help is much appreciated!
Best,
Pronabesh
Accepted Solutions
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Have you looked at the residuals in a plot? The ODS graphics in MIXED are pretty rich, and ought to give some hints. There is a possiblity that the residuals are multimodal, indicating that the variability differs by disease_severity. What happens if you fit:
proc mixed data=have;
title 'mixed model';
class subject day disease_severity;
model calorie=day|disease_severity; /* Changed to a factorial as the progression in time may differ by disease_severity */
repeated day/subject=subject type=un group=disease_severity; /* Fits separate covariances by disease_severity */
lsmeans day;
run;
If you have to move to a generalized model, a likely candidate for the distribution is lognormal, or possibly normal with a multiplicative error (link=log in GLIMMIX terms).
GLIMMIX only fits exponential family distributions, so that should not be a major concern.
Steve Denham
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Have you looked at the residuals in a plot? The ODS graphics in MIXED are pretty rich, and ought to give some hints. There is a possiblity that the residuals are multimodal, indicating that the variability differs by disease_severity. What happens if you fit:
proc mixed data=have;
title 'mixed model';
class subject day disease_severity;
model calorie=day|disease_severity; /* Changed to a factorial as the progression in time may differ by disease_severity */
repeated day/subject=subject type=un group=disease_severity; /* Fits separate covariances by disease_severity */
lsmeans day;
run;
If you have to move to a generalized model, a likely candidate for the distribution is lognormal, or possibly normal with a multiplicative error (link=log in GLIMMIX terms).
GLIMMIX only fits exponential family distributions, so that should not be a major concern.
Steve Denham
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Steve,
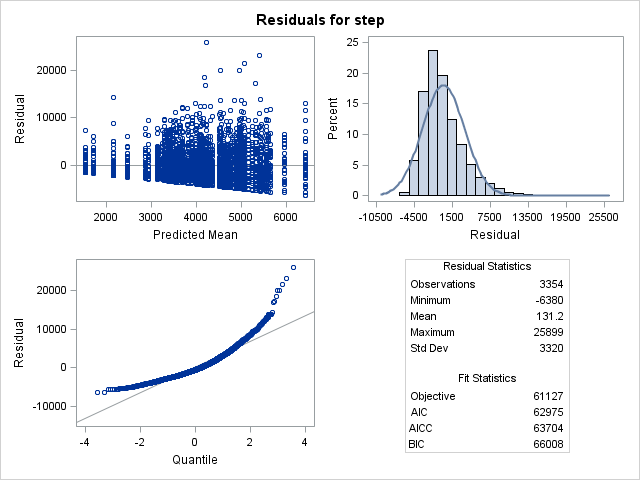
As always thank you for your advice. I used your suggested full factorial model with separate co-variances by disease_severity. The residual diagnostics is shown below. The distribution is negatively skewed as I expected.
I will also explore the fit statistics using two other approach:
1. Lognormal distribution as you suggested
2. Using multilevel model where disease_severity is a hierarchical level.
Note, the dependent variable here is step instead of calories, but the overall model is conceptually the same.
More to follow............
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
The histogram and residual plots scream log normal to me.
Steve Denham
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Thanks Steve.
I presume using a log normal distribution with link=log requires adding an ilink statement to the lsmeans to get the estimated marginal means. Is my assumption correct?
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
No. The ILINK for dist=lognormal retains values on the transformed scale, because you are modeling the log values as a normal random variable. To get estimates on the original scale, check out the documentation for the MODEL statement. DIST options, and you will see a section that starts out "When you choose DIST=LOGNORMAL..." There are equations for the expected value on the original scale, the variance on the original scale, and the everpopular omega. I use the Estimate value as mu and the squared StdErr value as sigma squared to get back to the original scale.
Try it and see... (sound like a drug pusher there).
Now for a model that assumes that the error is multiplicative (constant CV), dist=normal link=log, followed by ILINK in the lsmeans statement. Slightly different approach, and may be more appropriate to your data. This will return geometric means on the original scale.
Steve Denham
ANOVA, or Analysis Of Variance, is used to compare the averages or means of two or more populations to better understand how they differ. Watch this tutorial for more.
Find more tutorials on the SAS Users YouTube channel.



