- Home
- /
- Programming
- /
- Programming
- /
- reading fasta file into dataset
- RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
I have an input file with a sequence in FASTA format begins with a single-line description, followed by lines of sequence data.
The description line (defline) is distinguished from the sequence data by a greater-than (">") symbol at the beginning, shorter than 80 characters in length.
The data is divided into 50 character set each, in multiples lines extending upto 1400 characters.
>gi|5524211 gb AAD44166.1 cytochrome b
LCLYTHIGRNIYYGSYLYSETWNTGIMLLLITMATAFMGYVLPWGQMSFW
GATVITNLFSAIPYIGTNLVEWIWGGFSVDKATLNRFFAFHFILPFTMVA
LAGVHLTFLHETGSNNPLGLTSDSDKIPFHPYYTIKDFLGLLILILLLLL
LALLSPDMLGDPDNHMPADPLNTPLHIKPEWYFLFAYAILRSVPNKLGGV
LALFLSIVILGLMPFLHTSKHRSMMLRPLSQALFWTLTMDLLTLTWIGSQ
PVEYPYTIIGQMASILYFSIILAFLPIAGXIENY
My question: When I read the input file into a dataset, I created two columns, "Desc" and "Sequence". I need my dataset to have one Desc row and one Sequence row, but the sequence is getting divided up into multiple row like as follows. Looking for help either cleaning the LFCR as I create the dataset or conc the rows after the dataset is created. PLEASE HELP
Obs Desc Sequence
-------------------------------------------------------------------------------------------------------------------------
1 gi|5524211 gb AAD44166.1 cytochrome b LCLYTHIGRNIYYGSYLYSETWNTGIMLLLITMATAFMGYVLPWGQMSFW
2 GATVITNLFSAIPYIGTNLVEWIWGGFSVDKATLNRFFAFHFILPFTMVA
3 LAGVHLTFLHETGSNNPLGLTSDSDKIPFHPYYTIKDFLGLLILILLLLL
4 LALLSPDMLGDPDNHMPADPLNTPLHIKPEWYFLFAYAILRSVPNKLGGV
5 LALFLSIVILGLMPFLHTSKHRSMMLRPLSQALFWTLTMDLLTLTWIGSQ
6 PVEYPYTIIGQMASILYFSIILAFLPIAGXIENY
Accepted Solutions
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Ok. Now tested with Mike's (thanks Mike) fake data (with a single DESC) :
data want;
length desc $80 sequence $2000;
do until (eof);
infile "&sasforum.\datasets\fasta PG.txt" end=eof lrecl=1000;
input;
if char(_infile_,1) = '>' then desc = substr(_infile_,2);
else sequence = cats(sequence, _infile_);
end;
run;
PG
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
If, as you imply, there is only one desc per file, and thus, your dataset should contain only one observation, then this should do (untested) :
data want;
length desc $80 sequence $2000;
do until (eof);
infile "yourFastaFile.xxx" end=eof;
input;
if char(_infile_,1) = '>' then desc = substr(_infile_,2,80);
else sequence = cats(sequence, _infile_);
end;
run;
PG
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Thnx y'all, both responses works but the sequence is reading only upto 107 characters and not beyond. My input file has 1302 sequence char.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
hi ... can you post a portion of your data
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Ok. Now tested with Mike's (thanks Mike) fake data (with a single DESC) :
data want;
length desc $80 sequence $2000;
do until (eof);
infile "&sasforum.\datasets\fasta PG.txt" end=eof lrecl=1000;
input;
if char(_infile_,1) = '>' then desc = substr(_infile_,2);
else sequence = cats(sequence, _infile_);
end;
run;
PG
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
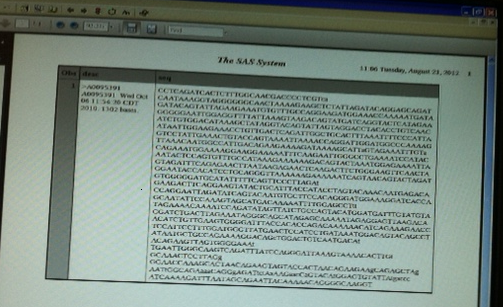
Thank you PGStats and MikeZdeb, your codes works perfectly as intended with Mike's and my dummy input file , but when I run it on my actual file (image below) it's not reading all the sequences using proc print.
I was not able to figure out why, Then It struck me maybe its something with my "proc print" output settings, so I used ODS to put in a pdf file, this time it read all my sequences but had spaces between the different sequences...hmm, Used ODS to html and boom all looks good (but cant explain why).
THANKYOU PGStats and MikeZdeb for your help and valuable time. Love this forum.
Input file

Incorrect output with spaces with ods pdf
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
hi ... if there are more than one DESC per file, I think this will work (at least it works with the attached fake data) ...
data slowa;
infile 'z:\fasta.txt' end=done;
length desc $100 sequence $1400;
do _n_=1 by 1 until (done);
input @;
if char(_infile_,1) eq '>' then do;
if _n_ ne 1 then output;
desc = substr(_infile_,2);
call missing(sequence);
end;
else sequence = cats(sequence,_infile_);
input;
end;
output;
run;
April 27 – 30 | Gaylord Texan | Grapevine, Texas
Registration is open
Walk in ready to learn. Walk out ready to deliver. This is the data and AI conference you can't afford to miss.
Register now and save with the early bird rate—just $795!
Learn how use the CAT functions in SAS to join values from multiple variables into a single value.
Find more tutorials on the SAS Users YouTube channel.
SAS Training: Just a Click Away
Ready to level-up your skills? Choose your own adventure.



