- Home
- /
- Programming
- /
- Programming
- /
- Update CSV with CRLF embedded
- RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
I found this code to remove embedded CRLF and update the file from SAS, but no update is taking place. No errors occur with this code. I have more than 300 csv files to import and about 10% of them have this issue. Some files have so many CRLF on the first row of data that import returns an error because it didn't detect any data on the first five rows of the file.
/************************** CAUTION ***************************/
/* */
/* This program UPDATES IN PLACE, create a backup copy before */
/* running. */
/* */
/************************** CAUTION ***************************/
/* Replace carriage return and linefeed characters inside */
/* double quotes with a specified character. This sample */
/* uses '@' and '$', but any character can be used, including */
/* spaces. CR/LFs not in double quotes will not be replaced. */
%let repA='@'; /* replacement character LF */
%let repD='$'; /* replacement character CR */
%let dsnnme="&folderbase.\Data\Raw\subfolder\filename.csv"; /* use full path of CSV file */
data _null_;
/* RECFM=N reads the file in binary format. The file consists */
/* of a stream of bytes with no record boundaries. SHAREBUFFERS */
/* specifies that the FILE statement and the INFILE statement */
/* share the same buffer. */
infile &dsnnme recfm=n sharebuffers;
file &dsnnme recfm=n;
/* OPEN is a flag variable used to determine if the CR/LF is within */
/* double quotes or not. Retain this value. */
retain open 0;
input a $char1.;
/* If the character is a double quote, set OPEN to its opposite value. */
if a = '"' then open = ^(open);
/* If the CR or LF is after an open double quote, replace the byte with */
/* the appropriate value. */
if open then do;
if a = '0D'x then put &repD;
else if a = '0A'x then put &repA;
end;
run;
*****************log when the code runs*****************************
NOTE: UNBUFFERED is the default with RECFM=N.
NOTE: The file/infile
"&folderbase.\Data\Raw\xxxx\xxxx.csv" is:
Filename=xxxxxx.csv,
RECFM=N,LRECL=256,File Size (bytes)=8530,
Last Modified=28Feb2023:14:33:14,
Create Time=28Feb2023:14:33:14
NOTE: UNBUFFERED is the default with RECFM=N.
NOTE: DATA statement used (Total process time):
real time 0.04 seconds
cpu time
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
If creating a .csv from an Excel sheet then normally the cells are comma delimited and the rows end with CRLF.
What you normally want to replace are the LF line feeds that come from a new line within a cell.
If that's the case you're dealing with then you could use some simpler code than the one you shared.
Do your .csv's have a header column on the first row and data starts on the second row? Or is the structure of higher complexity?
Ideally share one of your .csv's that have this problem here as an attachment (a few rows suffice, just change the data with a text editor in case there is something confidential in it).
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
The structure is something like the following:
Garbage row - - states the machine used to take the measurements
header row - this is incomplete and does not cover all the columns
Data row 1 - 1187 columns
Garbage row 2 - same as the first garbage row - give machine name
Data row 2 - 1187 columns
Sometimes there will be more rows of data but each one is separated by the "Garbage Row". The data rows and header row are semicolon delimited. I do not import the header row.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
@_Hopper wrote:
The structure is something like the following:
Garbage row - - states the machine used to take the measurements
header row - this is incomplete and does not cover all the columns
Data row 1 - 1187 columns
Garbage row 2 - same as the first garbage row - give machine name
Data row 2 - 1187 columns
Sometimes there will be more rows of data but each one is separated by the "Garbage Row". The data rows and header row are semicolon delimited. I do not import the header row.
Looks like you're getting somewhere with what Tom proposed.
If not then you really need to attach a .csv that contains a few lines including the header and the "garbage" lines so we can see and understand "reality".
Narrative is most of the time insufficient - like in yours it's not clear to me where the CR and LF sit and which one would need removal.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
@_Hopper wrote:
The structure is something like the following:
Garbage row - - states the machine used to take the measurements
header row - this is incomplete and does not cover all the columns
Data row 1 - 1187 columns
Garbage row 2 - same as the first garbage row - give machine name
Data row 2 - 1187 columns
Sometimes there will be more rows of data but each one is separated by the "Garbage Row". The data rows and header row are semicolon delimited. I do not import the header row.
If the pattern is that consistent then you probably do not need to modify the files. Just modify the code use to read the files. So if the 1187 "columns" are all numeric values then the program might be as simple as:
data want;
infile 'myfile' dsd dlm=';' truncover ;
input machine :$50.;
input v1-v1187;
run;- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
If the embedded CR and/or LF values are inside of quoted strings then you can use: %replace_crlf() to make a NEW file. You can use it to replace or remove either or both of those characters.
If they are not inside of quotes then you will have to figure out how to determine which line breaks should be removed. Perhaps by counting delimiters. Perhaps by detecting lines that start with a distinctive pattern.
If you have a lot of them then use a dataset with the list of files to generate one call to the macro per file.
For example if you have the data named FILES with a variable named FILENAME that has the names of the files (strings like 'case1.csv' and 'case2.csv'. And you want to write fixed files into a different directory using the same filename then the data step could look like:
data _null_;
set files;
length in out $255 ;
in=cats('/source/directory/name/',filename);
out=cats('/target/directory/name/',filename);
call execute(cats('%nrstr(%replace_crlr)(',quote(trim(in)),',',quote(trim(out)),')'));
run;- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
I have a macro setup to read all the files, so that's not an issue. I need to remove the hard carriage returns from each CSV then replace the file before importing to SAS.
This seems to work - testing it now. It does seem to run a bit slow but with 300+ files to process I suppose that's expected.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
This didn't appear to work either. Still have files not importing because of extra characters of some kind that SAS is misinterpreting.
The code runs fine, but there are still non-printable characters causing issues somewhere.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Log file:
NOTE: The infile
'xxx.csv' is:
Filename=A_zzz.csv,
RECFM=V,LRECL=32767,File Size (bytes)=8530,
Last Modified=31Aug2023:09:08:57,
Create Time=31Aug2023:09:08:45
NOTE: 1 record was read from the infile
'A_zzz.csv'.
The minimum record length was 3998.
The maximum record length was 3998.
NOTE: The data set WORK.OCTOEXP08 has 0 observations and 1158 variables.
NOTE: DATA statement used (Total process time):
real time 0.03 seconds
cpu time 0.03 seconds
0 rows created in WORK.OCTOEXP08 from
A_zzz.csv.
The SAS System
NOTE: WORK.OCTOEXP08 data set was successfully created.
NOTE: The data set WORK.OCTOEXP08 has 0 observations and 1158 variables.
NOTE: PROCEDURE IMPORT used (Total process time):
real time 0.19 seconds
cpu time 0.17 seconds
NOTE: PROCEDURE SQL used (Total process time):
real time 0.00 seconds
cpu time 0.00 seconds
WARNING: dataset A_zzz.csv did not import correctly
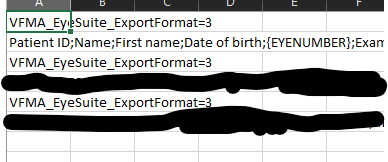
This is the suspect file - all have the same layout. The data rows are the ones blacked out.
When I go to the end of the first line and manually hit delete several times, this takes care of the issue. Not sure what characters are causing it because they are non-printable.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
If the file is only 8,530 bytes long just read in the whole file and LOOK at it to see where the CR ('0D'x) or LF ('0A'x) or other strange characters appear.
data _null_;
infile 'A_zzz.csv' recfm=f lrecl=100;
input;
list;
run;- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Ok - I see the following characters:
Garbage line 1: VFMA_EyeSuite_ExportFormat=3.PatientID;<-this is the header on the next row
Garbage line 2: VFMA_EyeSuite_ExportFormat=3.0000z;<---this also repeats on the third garbage line has a delimiter
When opening the file all I see is Format=3.
In the files that process correctly, this is what it looks like:
Garbage Line 1: VFMA_EyeSuite_ExportFormat=3..Patient ID;<-double period
Garbage Line 2: VFMA_EyeSuite_ExportFormat=3..0000z;<-same as the file that won't process
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
You need to read the HEX CODES that the LIST statement generated to see what characters those periods in the CHAR line actually are.
Here let's make an example so you can see how the LIST statement formats the output when the line has non-printable characters.
filename csv temp;
data _null_;
file csv termstr=crlf ;
put 'field1;field2';
put 'abc' '0D'x '123' '0A'x ;
put 'Tab' '09'x 'character;999' ;
run;So when I read that back in you can see that if puts the hexcodes under that characters on line 2 and 3. And it write the number of characters in the line after the end of the CHAR line.
697 data _null_;
698 infile csv termstr=crlf;
699 input;
700 list;
701 run;
NOTE: The infile CSV is:
Filename=C:\Users\ABERNA~1\AppData\Local\Temp\1\SAS Temporary Files\_TD14144_AMRAPY3WVP0VKU0_\#LN00057,
RECFM=V,LRECL=32767,File Size (bytes)=44,
Last Modified=31Aug2023:11:35:25,
Create Time=31Aug2023:11:35:25
RULE: ----+----1----+----2----+----3----+----4----+----5----+----6----+----7----+----8----+----9----+----0
1 field1;field2 13
2 CHAR abc.123. 8
ZONE 66603330
NUMR 123D123A
3 CHAR Tab.character;999 17
ZONE 56606667667673333
NUMR 4129381213452B999
NOTE: 3 records were read from the infile CSV.
The minimum record length was 8.
The maximum record length was 17.
NOTE: DATA statement used (Total process time):
real time 0.00 seconds
cpu time 0.00 seconds
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
If the file has data on every other line then just read it that way.
It looks like you only have a hand full of columns so you are not going to get much added value from trying to get PROC IMPORT to GUESS what variables there are. Just write your own data step to read the file.
Just skip over the header line and start reading at the first data line.
data want;
infile 'A_zzz.csv' dsd dlm=';' truncover firstobs=4 ;
input @;
if _infile_=: 'VFMA_EyeSuite' then delete;
input ID :$10. Name :$30. FName :$30. DOB :anydtdte. Eyenumber Exam ....;
format dob yymmdd10.;
run;- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
This method does not work either. Bombs out on the same file each time.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
@_Hopper wrote:
This method does not work either. Bombs out on the same file each time.
Without more information there is no way to help.
If it consistently fails on one file but works on others then it seems clear that the one file is different in some way.
Learn how use the CAT functions in SAS to join values from multiple variables into a single value.
Find more tutorials on the SAS Users YouTube channel.
SAS Training: Just a Click Away
Ready to level-up your skills? Choose your own adventure.