- Home
- /
- Programming
- /
- Programming
- /
- Re: The number of obs in datasets do not add up after MERGE
- RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
This might be a simple task but I could not figure it out. I am trying to merge 3 datasets together. if they dont merge by the variables, then split out to a nomatch dataset. Theoretically, the matches and no-matches number should add up to the number of obs in the datasets. Let's look at my code for better understanding
data MATCH
nomatch_A
nomatch_B
nomatch_C;
merge data_A(in=A)
data_B(in=B)
data_C(in=C);
by var1 var2 var3 var4;
if A and B and C then output MATCH;
else if A and (not B or not C) then output nomatch_A;
else if B and (not C or not A) then output nomatch_B;
else if C and (not A or not B) then output nomatch_C;
run;so if A has 1,000 obs and there are 800 obs in the dataset MATCH, then nomatch_A should have 200 obs. However, I got a number less than 200. Why? where did the other go?
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Hello,
I think there is a problem with the logic behind datasets nomatch_B and nomatch_C in that they are only populated
if the preceeding conditions are not matched.
For example, if an observation is in A and B but not in C, then it will not appear in nomatch_B.
Try (no data => not tested) :
if A and B and C then output MATCH;
else do;
if A then output nomatch_A;
if B then output nomatch_B;
if C then output nomatch_C;
end;
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
I would recommend adjusting how you think about this problem. Let's look at an approach first, and discuss a couple of the pitfalls later. Consider:
data want;
merge data_A(in=A)
data_B(in=B)
data_C(in=C);
by var1 var2 var3 var4;
match_pattern = '---';
if A then substr(match_pattern, 1, 1) = 'A';
if B then substr(match_pattern, 2, 1) = 'B';
if C then substr(match_pattern, 3, 1) = 'C';
run;
proc freq data=want;
tables match_pattern;
run;This creates a single variable that shows, for each observation, where it came from. That should be sufficient for you to determine what should happen next.
Some of the issues may involve your expectations of what the number of observations should be. To explore that, consider a simplified case where DATA_A and DATA_B both contain 10 observations, and we're merging by just one variable named ID.
data test1;
merge data_A data_B;
by ID;
run;The number of observations generated could be anywhere from 10 to 20, depending on the number of matches and mismatches. If you allow that either data set might contain duplicate values for the same ID, the complexity increases:
data test2;
merge data_A (in=A) data_B;
by ID;
if A;
run;Now the number of observations generated must be at least 10, but could be as many as 19. The 19 would occur of DATA_B contains just a single ID, repeated 10 times, that matches one of the IDs in DATA_A.
If both DATA_A and DATA_B could contain duplicate observations for the same ID, the problem gets so complex that SAS issues a warning in the log ... something along the lines of "more than one data set contains multiple observations for ID". So be sure to check the log for that type of message.
At any rate, the first program should at least give you a chance to figure this one out.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
BTW, this is one of the nice features of the SUBSTR function - it can be put on the left of the equals sign in an assignment statement. Just the ticket here.
The hash OUTPUT method will overwrite a SAS data set, but not append. That can be costly. Consider voting for Add a HASH object method which would append a hash object to an existing SAS data set
Would enabling PROC SORT to simultaneously output multiple datasets be useful? Then vote for
Allow PROC SORT to output multiple datasets
--------------------------
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
thank you for the detailed response. I have removed all duplicates so there would not be any obs with more than 1 match.
Your first set of code helps to see where the obs come from, but I don't think it show the ones that are missing (because the numbers don't add up)
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
I can't see the numbers that you are looking at. If you would like feedback ...
Post the number of observations you start with from each source (after you had removed duplicates), and the PROC FREQ results.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
I would suggest creating non-temporary variables with the values of the IN variables and you can examine explicitly which combinations of values are appearing "incorrectly" in which set and adjust logic accordingly.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
can you please explain a bit more on the "non-temporary variables"? what should I create?
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
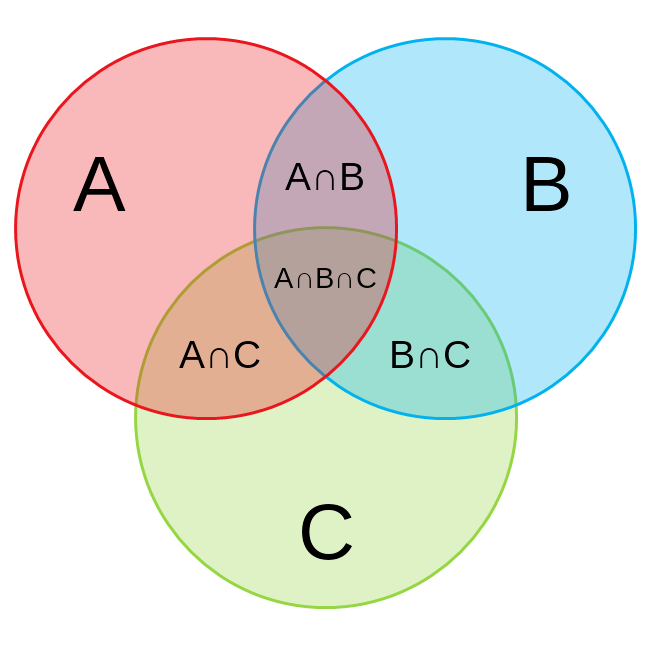
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
The answer lies in how you set up your clauses
if A and (not B or not C) translates to if A =1 and (B=0 or C=0)
possible outcomes
A=1 B=1 C=1
A=1 B=0 C=1
A=1 B=1 C=0
A=1 B=0 C=0
if your objective is to truly capture the fourth case then the condition becomes A=1 and (B=0 and C=0) and the counts will add up
The second and third conditions are captured by
if B and (Not A or Not C), if C and (Not A or Not B) so the datasets are not mutually exclusive and hence the row count does not add up.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
The are 8 possible ways to populate 3 binary variables. One combination is impossible (all false). Your program is only writing out 4. So either you skipped some of them or included them into multiple datasets.
Here is how to get all 7 combinations.
data
MATCH
A_ONLY
B_ONLY
C_ONLY
AB_ONLY
AC_ONLY
BC_ONLY
;
merge data_A(in=A) data_B(in=B) data_C(in=C);
by var1 var2 var3 var4;
if A and B and C then output MATCH;
else if A and not (B or C) then output A_ONLY;
else if B and not (C or A) then output B_ONLY;
else if C and not (A or B) then output C_ONLY;
else if A and B then output AB_ONLY;
else if A and C then output AC_ONLY;
else if B and C then output BC_ONLY;
else put 'This combination is impossible. ' (_n_ a b c) (=);
run;Checking your logic it seems you do account for all 7 combinations.
data test;
do a=1,0; do b=1,0; do c=1,0;
length m1 m2 $20;
if A and B and C then m1='MATCH';
else if A and (not B or not C) then m1='nomatch_A';
else if B and (not C or not A) then m1='nomatch_B';
else if C and (not A or not B) then m1='nomatch_C';
else m1='none';
if a and b and c then m2='MATCH';
else if a and b and not c then m2='AB_ONLY';
else if a and not b and c then m2='AC_ONLY';
else if not a and b and c then m2='BC_ONLY';
else if a then m2='A_ONLY';
else if b then m2='B_ONLY';
else if c then m2='C_ONLY';
else m2='none';
output;
end; end; end;
run;Not clear if this is exactly what you wanted.
Obs a b c m1 m2 1 1 1 1 MATCH MATCH 2 1 1 0 nomatch_A AB_ONLY 3 1 0 1 nomatch_A AC_ONLY 4 1 0 0 nomatch_A A_ONLY 5 0 1 1 nomatch_B BC_ONLY 6 0 1 0 nomatch_B B_ONLY 7 0 0 1 nomatch_C C_ONLY 8 0 0 0 none none
April 27 – 30 | Gaylord Texan | Grapevine, Texas
Registration is open
Walk in ready to learn. Walk out ready to deliver. This is the data and AI conference you can't afford to miss.
Register now and lock in 2025 pricing—just $495!
Learn how use the CAT functions in SAS to join values from multiple variables into a single value.
Find more tutorials on the SAS Users YouTube channel.
SAS Training: Just a Click Away
Ready to level-up your skills? Choose your own adventure.



