- Home
- /
- Programming
- /
- Programming
- /
- Re: Need help with correct counting with Proc SQL
- RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
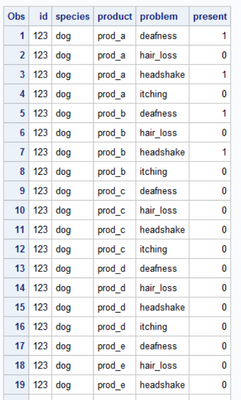
Dear experts,
I have a dataset containing columns for ID, species, product and problem. In the end I want to calculate the Odds Ratio for each pair of product and problem per species, meaning that I have to determine the values for a 2x2 crosstable for each row. Each ID can have multiple values for product and problem (but only one species). It is important, that each ID is counted only once in the crosstable.
I'm working with SAS 9.4. Here is an example dataset:
data raw;
input id species $ product $ problem $15-25;
datalines;
123 dog prod_a deafness
123 dog prod_a headshake
123 dog prod_b deafness
123 dog prod_b headshake
345 dog prod_a itching
345 dog prod_c itching
234 cat prod_c hair_loss
567 cat prod_d hair_loss
678 cat prod_e deafness
321 dog prod_a deafness
321 dog prod_a headshake
321 dog prod_c deafness
321 dog prod_c headshake
543 dog prod_a itching
543 dog prod_c itching
432 cat prod_c hair_loss
765 cat prod_d hair_loss
876 cat prod_e deafness
;
run;After sorting by species to enable species specific analyses, I want to count the IDs for my crosstable under the following conditions:
A = [IDs with the product and with the problem]
B = [IDs with the product but without the problem]
C = [IDs without the product but with the problem]
D = [IDs without the product and without the problem]
The ROR is then calculated with (A/B)/(C/D).
This is my code:
proc sql;
select distinct raw.species, raw.product, raw.problem,
(select count(id) from raw sub
where sub.species = raw.species
and sub.id ^= raw.id
and sub.product = raw.product
and sub.problem = raw.problem) as a,
(select count(id) from ror sub
where sub.species = raw.species
and sub.id ^= raw.id
and sub.product = raw.product
and sub.problem ^= raw.problem) as b,
(select count(id) from raw sub
where sub.species = raw.species
and sub.id ^= raw.id
and sub.product ^= raw.product
and sub.problem = raw.problem) as c,
(select count(id) from raw sub
where sub.species = raw.species
and sub.id ^= raw.id
and sub.product ^= raw.product
and sub.problem ^= raw.problem) as d,
(calculated a/ calculated b)/(calculated c/calculated d) as ror
from raw;
quit;This is the result:
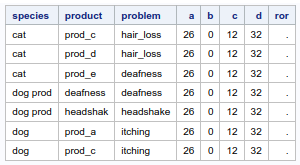
Now I have two problems:
- „a“ is expected to be at least 1, but the code does not count the ID itself (that's logic, as it counts where sub.id ^= raw.id)
- From the preliminary results, I can see, that with this code each ID can be counted more than once (thus contributing to more than one field of the 2x2 table). This must not be the case.
Is there anyone out there, who can help with the solution? I hope I made myself clear enough...
Looking forward to your hints and ideas!
Kind regards,
SabineT
Accepted Solutions
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
You seem to be trying to make a 2x2 table. If you do a proc summary with two class variables and request all of the possible combinations SAS will create a _TYPE_ variable that goes from 0 to 3 with 0 meaning the overall summary, 1 being the summary by just the second class variable, 2 being the summary be just the first class variable and then 3 being the summary by both class variables. You can also think of N1 and N2 as being the row and column sum and N0 as the overall sum.
So if we treat those four summaries as N0 to N3 the table below shows the relationship between your A,B,C,D labels for the cells and how to calculate them based on the summaries.
| V-Product Problem---> | Present | Absent | Total |
| Present | A =N3 | B=N1-N3 | N1 |
| Absent | C = N2-N3 | D = N0-N1-N2+N3 | N0-N2 |
| Total | N2 | N0-N2 | N0 |
So given your example data:
data raw;
input species $ product $ problem :$20. id;
datalines;
cat prod_a vomit 666
cat prod_c deafness 777
cat prod_c hair_loss 234
cat prod_c hair_loss 432
cat prod_c vomit 666
cat prod_d hair_loss 567
cat prod_d hair_loss 765
cat prod_e deafness 678
cat prod_e deafness 876
cat prod_e hair_loss 999
cat prod_e vomit 999
cat prod_g hair_loss 888
dog prod_a deafness 123
dog prod_a deafness 321
dog prod_a headshake 123
dog prod_a headshake 321
dog prod_a itching 345
dog prod_a itching 543
dog prod_a vomit 333
dog prod_b deafness 123
dog prod_b headshake 123
dog prod_c deafness 321
dog prod_c headshake 321
dog prod_c itching 345
dog prod_c itching 543
dog prod_c vomit 444
dog prod_f hair_loss 111
dog prod_g hair_loss 222
dog prod_g hair_loss 555
dog prod_h hair_loss 555
;We can create datasets with the N0, N1, N2 and N3 variables this way with SQL.
proc sql;
create table n0 as select species,count(distinct id) as n0 from raw group by 1;
create table n1 as select species,product,count(distinct id) as n1 from raw group by 1,2 ;
create table n2 as select species,problem,count(distinct id) as n2 from raw group by 1,2 ;
create table n3 as select a.species,a.product,a.problem,sum(not missing(b.id)) as n3
from (select species,product,problem,id
from (select distinct id,species from raw)
, (select distinct product from n1)
, (select distinct problem from n2)
) a
natural left join raw b
group by 1,2,3
;And you can then combine them and also calculate A, B, C, D and ROR this way:
create table summary as
select *
, n3 as A
, n1-n3 as B
, n2-n3 as C
, n0-n1-n2+n3 as D
,(n3/(n1-n3))/((n2-n3)/(n0-n1-n2+n3)) as ROR
from n0 natural join n1 natural join n2 natural join n3
;Results:
Obs species product problem n3 n0 n1 n2 A B C D ROR
1 cat prod_a deafness 0 10 1 3 0 1 3 6 0.0000
2 cat prod_a hair_loss 0 10 1 6 0 1 6 3 0.0000
3 cat prod_a vomit 1 10 1 2 1 0 1 8 .
4 cat prod_c deafness 1 10 4 3 1 3 2 4 0.6667
5 cat prod_c hair_loss 2 10 4 6 2 2 4 2 0.5000
6 cat prod_c vomit 1 10 4 2 1 3 1 5 1.6667
7 cat prod_d deafness 0 10 2 3 0 2 3 5 0.0000
8 cat prod_d hair_loss 2 10 2 6 2 0 4 4 .
9 cat prod_d vomit 0 10 2 2 0 2 2 6 0.0000
10 cat prod_e deafness 2 10 3 3 2 1 1 6 12.0000
11 cat prod_e hair_loss 1 10 3 6 1 2 5 2 0.2000
12 cat prod_e vomit 1 10 3 2 1 2 1 6 3.0000
13 cat prod_g deafness 0 10 1 3 0 1 3 6 0.0000
14 cat prod_g hair_loss 1 10 1 6 1 0 5 4 .
15 cat prod_g vomit 0 10 1 2 0 1 2 7 0.0000
16 dog prod_a deafness 2 9 5 2 2 3 0 4 .
17 dog prod_a hair_loss 0 9 5 3 0 5 3 1 0.0000
18 dog prod_a headshake 2 9 5 2 2 3 0 4 .
19 dog prod_a itching 2 9 5 2 2 3 0 4 .
20 dog prod_a vomit 1 9 5 2 1 4 1 3 0.7500
21 dog prod_b deafness 1 9 1 2 1 0 1 7 .
22 dog prod_b hair_loss 0 9 1 3 0 1 3 5 0.0000
23 dog prod_b headshake 1 9 1 2 1 0 1 7 .
24 dog prod_b itching 0 9 1 2 0 1 2 6 0.0000
25 dog prod_b vomit 0 9 1 2 0 1 2 6 0.0000
26 dog prod_c deafness 1 9 4 2 1 3 1 4 1.3333
27 dog prod_c hair_loss 0 9 4 3 0 4 3 2 0.0000
28 dog prod_c headshake 1 9 4 2 1 3 1 4 1.3333
29 dog prod_c itching 2 9 4 2 2 2 0 5 .
30 dog prod_c vomit 1 9 4 2 1 3 1 4 1.3333
31 dog prod_f deafness 0 9 1 2 0 1 2 6 0.0000
32 dog prod_f hair_loss 1 9 1 3 1 0 2 6 .
33 dog prod_f headshake 0 9 1 2 0 1 2 6 0.0000
34 dog prod_f itching 0 9 1 2 0 1 2 6 0.0000
35 dog prod_f vomit 0 9 1 2 0 1 2 6 0.0000
36 dog prod_g deafness 0 9 2 2 0 2 2 5 0.0000
37 dog prod_g hair_loss 2 9 2 3 2 0 1 6 .
38 dog prod_g headshake 0 9 2 2 0 2 2 5 0.0000
39 dog prod_g itching 0 9 2 2 0 2 2 5 0.0000
40 dog prod_g vomit 0 9 2 2 0 2 2 5 0.0000
41 dog prod_h deafness 0 9 1 2 0 1 2 6 0.0000
42 dog prod_h hair_loss 1 9 1 3 1 0 2 6 .
43 dog prod_h headshake 0 9 1 2 0 1 2 6 0.0000
44 dog prod_h itching 0 9 1 2 0 1 2 6 0.0000
45 dog prod_h vomit 0 9 1 2 0 1 2 6 0.0000
If you don't want all of the possible combinations you could limit N3 to just the actual combinations that appear.
That is easier to calculate:
create table n3 as select species,product,problem,count(distinct id) as n3 from raw group by 1,2,3 ;
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
I'm kind of lost by your definitions of A B C D.
I feel as if you are working very hard to do something relatively simple, but as I'm not sure what you are trying to do, I can't really say what the simple solution is.
Let me guess what you want ... do you want a table which has all possible combinations of species, product and problem, and associated counts? Am I close, or way off?
Paige Miller
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Hi PaigeMiller,
many thanks for taking care!
I even thought that my task is quite simple, but I went a lot of ways and this time the output appears close to the target.
What I want to calculate is the frequency of each pair of product and problem (drug-event-pair -> DEP) per species. The ratio (ROR) should tell me if the frequency is higher than expected (>1). This means a potential signal in pharmacovigilance (= monitoring the benefit-risk profile of drugs). In the very end I am not interested in the table but only in the ROR.
A is the number of DEPs of interest. B is the number of events with the same product but without the problem. C is the number of events with the same problem but without the drug of interest. And under D all events should be counted that do neither contain the event nor the drug of interest.
I think the problem is that SAS checks the rows and I have to find a way that it checks all rows for one ID to assign the ID to the right field of the crosstable.
I hope that helps? 🙄
Regards, Sabine
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
@SabineT wrote:
A is the number of DEPs of interest. B is the number of events with the same product but without the problem. C is the number of events with the same problem but without the drug of interest. And under D all events should be counted that do neither contain the event nor the drug of interest.🙄 I hope that helps?
It doesn't really help, when I say I don't understand what A B C D is, to repeat the definition with different words. And I don't know what "DEPs" means. I want more explanation. I want examples. Walk me through how A is calculated in the sample data provided. Walk me through how B and C and D is calculated in the sample data provided.
By the way, in general, when code isn't giving you the right answer, we need to know what the right answer is.
Paige Miller
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Sorry, I tried to keep it short - but obviously not simple...
With the sample data I would expected the following output:
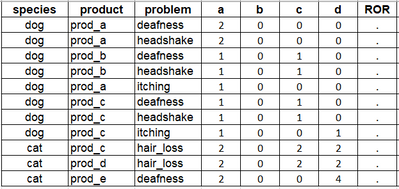
I should have included any reports with "new" symptoms to have some counts for B... 🤔
For car I counted the IDs that contain prod_c and hair_loss as 'a' for that drug event pair. It is 234 and 432, so 2 IDs. Then I look for any other ID containing prod_c but not hair_loss, but there i none (b=0). For 'c' I count those IDs that do not have prod_c but hair_loss. 567 and 765 satisfy this condition, so c = 2. 'd' is the count of IDs that contain neither prod_c nor hair_loss. Here we have 678 and 876 (d=2). ROR can't be calculated since 0 is included as denominator.
I listed the expected counts in the following table:
| species | product | problem | a | IDs | b | IDs | c | IDs | d | IDs |
| dog | prod_a | deafness | 2 | 123, 321 | 0 | 0 | 0 | |||
| dog | prod_a | headshake | 2 | 123, 321 | 0 | 0 | 0 | |||
| dog | prod_b | deafness | 1 | 123 | 0 | 1 | 321 | 0 | ||
| dog | prod_b | headshake | 1 | 123 | 0 | 1 | 321 | 0 | ||
| dog | prod_a | itching | 1 | 321 | 0 | 0 | 0 | |||
| dog | prod_c | deafness | 1 | 321 | 0 | 1 | 123 | 0 | ||
| dog | prod_c | headshake | 1 | 321 | 0 | 1 | 123 | 0 | ||
| dog | prod_c | itching | 1 | 321 | 0 | 0 | 1 | 123 | ||
| cat | prod_c | hair_loss | 2 | 234, 432 | 0 | 2 | 567, 765 | 2 | 678, 876 | |
| cat | prod_d | hair_loss | 2 | 567, 765 | 0 | 2 | 234, 432 | 2 | 678, 876 | |
| cat | prod_e | deafness | 2 | 678, 876 | 0 | 0 | 4 | 234, 432, 567, 765 |
You see the sum of A to D should always be the sum of IDs per species.
And as I describe the steps to walk through the dataset, I understand that the conditions should not be simply connected with AND in the code...
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
I suspect that you are using the letters A,B,C,D to refer to the cells in a 2 by 2 table.
data example;
input treated sick label $;
cards;
1 1 A
1 0 B
0 1 C
0 0 D
;So to get that 2x2 table why not just convert the way you have the data so that you have these two binary variables?
It is hard to get the negative cells from the way you stored the data. Can you explain the logic of how to calculate them?
So for a particular ID to you want to use it for all of the possible products? Or only the products that it ever received? Similarly do you want to count that ID for every possible disease? Or only the diseases that you have included for it for some specific product?
If you want to generate 0/1 flags for every possible combination you could do something like this:
data raw;
input id species :$10. product :$10. problem :$20. ;
datalines;
123 dog prod_a deafness
123 dog prod_a headshake
123 dog prod_b deafness
123 dog prod_b headshake
345 dog prod_a itching
345 dog prod_c itching
234 cat prod_c hair_loss
567 cat prod_d hair_loss
678 cat prod_e deafness
321 dog prod_a deafness
321 dog prod_a headshake
321 dog prod_c deafness
321 dog prod_c headshake
543 dog prod_a itching
543 dog prod_c itching
432 cat prod_c hair_loss
765 cat prod_d hair_loss
876 cat prod_e deafness
;
proc sort data=raw ;
by id species product problem ;
run;
proc sql ;
create table skeleton as
select *
from (select distinct id,species from raw)
, (select distinct product from raw)
, (select distinct problem from raw)
order by id, species, product, problem
;
quit;
data want ;
merge raw (in=in1) skeleton;
by id species product problem ;
present=in1;
run;So that you get a table something like this:
From which it should be easier to calculate your A,B,C,D numbers.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
This is helpful, but I'm still not there.
Questions:
- Why does dog/prod_a/itching have only 1 for A? I see two dog/prod_a/itching in the raw data.
- What would cause B to be greater than zero? If there are no examples of this in the data, could you modify the data so there is an example or two where B>0? Otherwise, I can't program it.
- What does this mean? "It is important, that each ID is counted only once in the crosstable." When an ID appears more than once, which of the multiple records for that ID are not used?
Paige Miller
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
2. I will add some data contributing to B.
3. I stated this because SAS currently counts each ID more than once. But if I'm not on the wrong track, each ID falls in one of the categories A, B, C or D only.
>> So I will rework on my sample data and the expected output.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Hello everyone,
I hope you stay tuned 😅
Here is the optimised dataset:
data raw;
input id species $ product $ problem $15-25;
datalines;
123 dog prod_a deafness
123 dog prod_a headshake
123 dog prod_b deafness
123 dog prod_b headshake
345 dog prod_a itching
345 dog prod_c itching
234 cat prod_c hair_loss
567 cat prod_d hair_loss
678 cat prod_e deafness
321 dog prod_a deafness
321 dog prod_a headshake
321 dog prod_c deafness
321 dog prod_c headshake
543 dog prod_a itching
543 dog prod_c itching
432 cat prod_c hair_loss
765 cat prod_d hair_loss
876 cat prod_e deafness
111 dog prod_f hair_loss
222 dog prod_g hair_loss
333 dog prod_a vomit
444 dog prod_c vomit
555 dog prod_g hair_loss
555 dog prod_h hair_loss
666 cat prod_a vomit
666 cat prod_c vomit
777 cat prod_c deafness
888 cat prod_g hair_loss
999 cat prod_e vomit
999 cat prod_e hair_loss
;
run;the expected output:

(that is telling me - among other things - that the odds for deafness with prod_e are 12fold higher than for other prod)
I skipped the list of contributing IDs as it was postflooding...
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Did I scare you off!?
I tried to abstract my requirement as follows:
for each row do A = 1
B = 0
C = 0
D = 0 check all rows for each id together
different id? yes: does id contain the same species? > yes: does id contain the same product? > yes: does id contain the same problem? > yes: A + 1 no: B + 1 no: does id contain the same problem? > yes: C + 1 no: D + 1 no: continue with the next id
no: continue with the next row
Who is confident that there is a solution for this in SAS?
I am lost in reading threads about arrays and loops 😓
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
You seem to be trying to make a 2x2 table. If you do a proc summary with two class variables and request all of the possible combinations SAS will create a _TYPE_ variable that goes from 0 to 3 with 0 meaning the overall summary, 1 being the summary by just the second class variable, 2 being the summary be just the first class variable and then 3 being the summary by both class variables. You can also think of N1 and N2 as being the row and column sum and N0 as the overall sum.
So if we treat those four summaries as N0 to N3 the table below shows the relationship between your A,B,C,D labels for the cells and how to calculate them based on the summaries.
| V-Product Problem---> | Present | Absent | Total |
| Present | A =N3 | B=N1-N3 | N1 |
| Absent | C = N2-N3 | D = N0-N1-N2+N3 | N0-N2 |
| Total | N2 | N0-N2 | N0 |
So given your example data:
data raw;
input species $ product $ problem :$20. id;
datalines;
cat prod_a vomit 666
cat prod_c deafness 777
cat prod_c hair_loss 234
cat prod_c hair_loss 432
cat prod_c vomit 666
cat prod_d hair_loss 567
cat prod_d hair_loss 765
cat prod_e deafness 678
cat prod_e deafness 876
cat prod_e hair_loss 999
cat prod_e vomit 999
cat prod_g hair_loss 888
dog prod_a deafness 123
dog prod_a deafness 321
dog prod_a headshake 123
dog prod_a headshake 321
dog prod_a itching 345
dog prod_a itching 543
dog prod_a vomit 333
dog prod_b deafness 123
dog prod_b headshake 123
dog prod_c deafness 321
dog prod_c headshake 321
dog prod_c itching 345
dog prod_c itching 543
dog prod_c vomit 444
dog prod_f hair_loss 111
dog prod_g hair_loss 222
dog prod_g hair_loss 555
dog prod_h hair_loss 555
;We can create datasets with the N0, N1, N2 and N3 variables this way with SQL.
proc sql;
create table n0 as select species,count(distinct id) as n0 from raw group by 1;
create table n1 as select species,product,count(distinct id) as n1 from raw group by 1,2 ;
create table n2 as select species,problem,count(distinct id) as n2 from raw group by 1,2 ;
create table n3 as select a.species,a.product,a.problem,sum(not missing(b.id)) as n3
from (select species,product,problem,id
from (select distinct id,species from raw)
, (select distinct product from n1)
, (select distinct problem from n2)
) a
natural left join raw b
group by 1,2,3
;And you can then combine them and also calculate A, B, C, D and ROR this way:
create table summary as
select *
, n3 as A
, n1-n3 as B
, n2-n3 as C
, n0-n1-n2+n3 as D
,(n3/(n1-n3))/((n2-n3)/(n0-n1-n2+n3)) as ROR
from n0 natural join n1 natural join n2 natural join n3
;Results:
Obs species product problem n3 n0 n1 n2 A B C D ROR
1 cat prod_a deafness 0 10 1 3 0 1 3 6 0.0000
2 cat prod_a hair_loss 0 10 1 6 0 1 6 3 0.0000
3 cat prod_a vomit 1 10 1 2 1 0 1 8 .
4 cat prod_c deafness 1 10 4 3 1 3 2 4 0.6667
5 cat prod_c hair_loss 2 10 4 6 2 2 4 2 0.5000
6 cat prod_c vomit 1 10 4 2 1 3 1 5 1.6667
7 cat prod_d deafness 0 10 2 3 0 2 3 5 0.0000
8 cat prod_d hair_loss 2 10 2 6 2 0 4 4 .
9 cat prod_d vomit 0 10 2 2 0 2 2 6 0.0000
10 cat prod_e deafness 2 10 3 3 2 1 1 6 12.0000
11 cat prod_e hair_loss 1 10 3 6 1 2 5 2 0.2000
12 cat prod_e vomit 1 10 3 2 1 2 1 6 3.0000
13 cat prod_g deafness 0 10 1 3 0 1 3 6 0.0000
14 cat prod_g hair_loss 1 10 1 6 1 0 5 4 .
15 cat prod_g vomit 0 10 1 2 0 1 2 7 0.0000
16 dog prod_a deafness 2 9 5 2 2 3 0 4 .
17 dog prod_a hair_loss 0 9 5 3 0 5 3 1 0.0000
18 dog prod_a headshake 2 9 5 2 2 3 0 4 .
19 dog prod_a itching 2 9 5 2 2 3 0 4 .
20 dog prod_a vomit 1 9 5 2 1 4 1 3 0.7500
21 dog prod_b deafness 1 9 1 2 1 0 1 7 .
22 dog prod_b hair_loss 0 9 1 3 0 1 3 5 0.0000
23 dog prod_b headshake 1 9 1 2 1 0 1 7 .
24 dog prod_b itching 0 9 1 2 0 1 2 6 0.0000
25 dog prod_b vomit 0 9 1 2 0 1 2 6 0.0000
26 dog prod_c deafness 1 9 4 2 1 3 1 4 1.3333
27 dog prod_c hair_loss 0 9 4 3 0 4 3 2 0.0000
28 dog prod_c headshake 1 9 4 2 1 3 1 4 1.3333
29 dog prod_c itching 2 9 4 2 2 2 0 5 .
30 dog prod_c vomit 1 9 4 2 1 3 1 4 1.3333
31 dog prod_f deafness 0 9 1 2 0 1 2 6 0.0000
32 dog prod_f hair_loss 1 9 1 3 1 0 2 6 .
33 dog prod_f headshake 0 9 1 2 0 1 2 6 0.0000
34 dog prod_f itching 0 9 1 2 0 1 2 6 0.0000
35 dog prod_f vomit 0 9 1 2 0 1 2 6 0.0000
36 dog prod_g deafness 0 9 2 2 0 2 2 5 0.0000
37 dog prod_g hair_loss 2 9 2 3 2 0 1 6 .
38 dog prod_g headshake 0 9 2 2 0 2 2 5 0.0000
39 dog prod_g itching 0 9 2 2 0 2 2 5 0.0000
40 dog prod_g vomit 0 9 2 2 0 2 2 5 0.0000
41 dog prod_h deafness 0 9 1 2 0 1 2 6 0.0000
42 dog prod_h hair_loss 1 9 1 3 1 0 2 6 .
43 dog prod_h headshake 0 9 1 2 0 1 2 6 0.0000
44 dog prod_h itching 0 9 1 2 0 1 2 6 0.0000
45 dog prod_h vomit 0 9 1 2 0 1 2 6 0.0000
If you don't want all of the possible combinations you could limit N3 to just the actual combinations that appear.
That is easier to calculate:
create table n3 as select species,product,problem,count(distinct id) as n3 from raw group by 1,2,3 ;
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
you made my day! Without your explanations it would be pure magic... With your explanations (especially on N0-N3) I feel that I will fully understand your code as soon as I am familiar with the SQL join function.
Many thanks!!!
Sabine
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Hello @SabineT
As you pointed out correctly there is an issue with your SQL Code - the way you have performed the self join.
I am posting the updated SQL Code.
proc sql;
select distinct raw.species, raw.product, raw.problem,
(select count(r.id) from raw r, raw s
where s.species = r.species
and s.id ^= r.id
and s.product = r.product
and s.problem = r.problem) as a ,
(select count(r.id) from raw r, raw s
where s.species = r.species
and s.id ^= r.id
and s.product = r.product
and s.problem ^= r.problem) as b,
(select count(r.id) from raw r, raw s
where s.species = r.species
and s.id ^= r.id
and s.product ^= r.product
and s.problem = r.problem) as c,
(select count(r.id) from raw r, raw s
where s.species = r.species
and s.id ^= r.id
and s.product ^= r.product
and s.problem ^= r.problem) as d,
(calculated a/ calculated b)/(calculated c/calculated d) as ror
from raw;
quit;The result is as follows
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
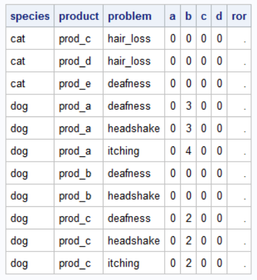
Hi Sajid01,
Thanks a lot for the helping step! Now the code is counting. But the result remains weired. How can D be 68 if there are only 18 observations? I'm posting the output again as it appears that the format crashed in your output:
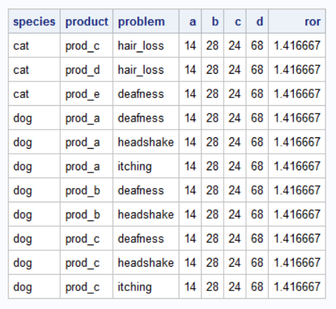
As posted to PaigeMiller the problem is that I don`t want to count observations but IDs (per species) depending on the conditions product and problem. It appears that SAS checks now the rows and counts the row once for each satisfied condition. So I am afraid, there is a very relevant step missing... Do you have an idea?
Regards, Sabine
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
We are doing a self join. It is a sort of Cartesian join (18 x 18=324) minus the filter conditions.
Please confirm if the logic in SQL is correct. I have used your logic and made only a minor correction to the SQL.
The data is also what you have provided.
Learn how use the CAT functions in SAS to join values from multiple variables into a single value.
Find more tutorials on the SAS Users YouTube channel.
SAS Training: Just a Click Away
Ready to level-up your skills? Choose your own adventure.