- Home
- /
- Programming
- /
- Programming
- /
- Re: Extract specific data from a variable with varying strings
- RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Hi,
I need to extract data from a variable in a data set that has notes about lab test results and test information. I'm trying to create 4 new columns with organism names (list of values with genus species names), gene mechanisms (KPC, NDM, VIM, OXA values), carbapenem-resistant (yes, no,unk values), and carbapenemase-producing(yes, no,unk values). Here are some examples of the Notes below. I highlighted the data that should be extracted for these notes into the new columns
- SPECIMEN NOTES:Collection Method ID: TMOT - Transport MediaNTE:75,000--99,000 cfu/ml Citrobacter freundii ***This CRE isolate demonstrates carbapenemase-producing - Here Citrobacter freundii should be extracted into the organism column and carbapenemase-producing as a yes to the carbapenemase-producing column.
- SPECIMEN NOTES:119334006 Sputum specimen SCT SPU SPUTUM L 20130731 v1NTE:=4+ CARBAPENEM RESISTANT ESCHERICHIA COLIOther - ESCHERICHIA COLI to organism and carbapenem resistant as a yes to carbapenem- resistant
- SPECIMEN NOTES: URC Urine clean catch HL70487 2.5 Urine clean catch
NTE: INTERPRETATION: A "Not Detected" Xpert? Carba-R Assay result does not preclude the presence of other resistance mechanisms. An ?Inconclusive? result cannot determine the presence or absence of blaIMP, blaVIM, blaNDM, blaKPC, and blaOXA-48 gene sequences. TEST DISCLAIMER: The Xpert? Carba-R Assay is a qualitative in vitro diagnostic test for the detection of blaKPC, blaNDM, blaVIM, blaOXA-48, and blaIMP gene sequences associated with carbapenem non-susceptible pure colonies of Enterobacteriaceae, Acinetobacter baumannii, or Pseudomonas aeruginosa. The Xpert? Carba-R Assay is not intended to guide or monitor treatment for carbapenem-non-susceptible bacterial infections. The performance of this assay was validated by the Microbial Diseases Laboratory. - This is all informational. No extraction - NTE:=2+ Klebsiella pneumoniae Carbapenemase-producing Enterobacteriaceae Positivefor Carbapenemase: KPC COLISTIN 2 INTERMEDIATE- Klebsiella pneumoniae to organism column and KPC to Gene-mechanism
- SPECIMEN NOTES:Collection Method ID: TMOT - Transport MediaNTE:70,000 cfu/ml Citrobacter brakii CRE (carbapenem-resistant Enterobacteriaceae) Targeted carbapenemase producing genes NOT detectedOther-Citrobacter brakii to organism, yes to carbapenem-resistant and no to carbapenemase-producing
I tried to replace each note with the data needed but this was not really an efficient idea xince there were about 1300 different notes among a 5600 dataset.
I know this is so complicated but I'm wondering if there is a way to work on this.
Any help would be really appreciated.
Thanks in advance
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Which values go into which new variable(s)?
What are the rules involved? You provide a very small list of examples. If those are not the only things you are looking for then you need to provide rules of exactly what you are looking for, plus rules for what variable(s) get what value based on finding them.
If the list of items you look for is relatively small then it might be possible to code something using FINDW function in a manner that ignores letter case. That would indicate that you need to provide a complete list of what you are searching for and how each is indicated as found in your data set.
Note that even your example has issues with spelling other than case CARBAPENEM RESISTANT and carbapenem-resistant, inconsistent use of hyphen.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
I can attach an Excel worksheet, maybe it would be clearer that way but I do not know how to attach the file. Is there any website or document to help me in attaching the file?
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
@mayasak wrote:
I can attach an Excel worksheet, maybe it would be clearer that way but I do not know how to attach the file. Is there any website or document to help me in attaching the file?
You are talking about text strings. No need to attach a binary formatted file to share simple text strings. Just paste the actual text into one of the text boxes opened when you click on the Insert Code or Insert SAS Code buttons on the forum editor form.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Hi Ballard,
There are 4 variables:
- Organism: I have a list of around 20 values that falls into this variable.
- Gene_Mechanism: 5 values (KPC, NDM, VIM, OXA, No gene), Anyone in notes will fill in the variable.
- Carbapenem_Resistant: If the string has carbapenem-resistant detected or carbapenem-resistant (with or without hyphen) positive, Yes would be filled in the variable. Any carbapenem resistant (or carbapenem-resistant) not detected or negative, No will fill in the variable, if no mention of carbapenem resistant (carbapenem-resistant), Unk will fill in the variable.
- Carbapenemase _Producing: Same as carbapenem resistant but for carbabenem producing.
Thank you
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
I cannot make any sense of that description.
Post a simple example of what you have using a data step.
For example to make a dataset with one observation and one variable write code like this:
data have;
length key $200;
key='xxxxx';
run;To make a dataset with multi observations and multiple variables then use something like this instead.
data have;
length var1 $100 var2 $40 ;
infile datalines dsd truncover ;
input var1 var2;
datatlines4;
Organism,Gen_mechanism
Carbpenem,Resitance
;;;;Do the same thing to demonstrate what OUTPUT dataset you want to get for that given input.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Hi Tom,
This is an example of what is needed below:
data have;
length Notes $256;
infile datalines dsd truncover;
input Notes $;
datalines4;
NTE:=2+ Klebsiella pneumoniae Carbapenemase-producing Enterobacteriaceae Positivefor Carbapenemase: NDM COLISTIN 2 INTERMEDIATE- Klebsiella pneumoniae to organism column and NDM to Gene-mechanism
SPECIMEN NOTES:Collection Method NTE:75 --99 cfu/ml Citrobacter freundii This CRE isolate demonstrates carbapenemase-producing
SPECIMEN NOTES:119334006 Sputum specimen SPUTUM 1NTE:=4+ CARBAPENEM RESISTANT ESCHERICHIA COLI
NTE:=2+ Klebsiella pneumoniae Carbapenemase-producing Enterobacteriaceae Positivefor Carbapenemase: KPC COLISTIN 2 INTERMEDIATE- Klebsiella pneumoniae to organism column and KPC to Gene-mechanism
SPECIMEN NOTES:Collection Method ID: TMOT - Transport MediaNTE:70 cfu/ml Citrobacter brakii CRE (carbapenem-resistant Enterobacteriaceae) Targeted carbapenemase producing genes NOT detectedOther-Citrobacter brakii to organism, yes to carbapenem-resistant and no to carbapenemase-producing
NTE:INTERPRETATION: An ?Inconclusive? result cannot determine the presence or absence of blaVIM blaNDM blaKPC and blaOXA-48 associated with carbapenem non-susceptible Acineto baumannii, or Pseudo aeruginosa.
SPECIMEN NOTES:Collection Method ID: CANTE:Candida aurisOther Identifiers: PI: 14310303980 PI: CCRFFLO5JXTLH
SPECIMEN NOTES:Collection Method ID: TMOT - Transport MediaNTE:Recovered Klebsiella aerogenes ***This CRE isolate demonstrates carbapenemase production.** Positive for KPC gene for carbapenemase production by PCR.Carbapenem resistant organism.
SPECIMEN NOTES:119334006 Sputum specimen SCT SPU SPUTUM L 20130731 v1NTE:=1+ Carbapenem resistant Klebsiella pneumonaiae POSITIVE Class A CarbapenemaseProducerOther Identifiers: CCREXRZYLDMRP
SPECIMEN NOTES:Collection Method: TRACHNTE:Carbapenem-resistant Enterobacteriaceae (CRE) - Klebsiella oxytocaSusceptibility profile is consistent with a probable ESBL.Carbapenemase not detected
;;;;
run;
data want8;
infile datalines delimiter=",";
length Organism $25 Gene_Mechanism $6 Carbapenem_Resistance $6 Carbapenemase_Producing $6;
input Organism $ Gene_Mechanism $ Carbapenem_Resistance $ Carbapenemase_Producing $;
datalines;
Klebsiella aerogenes,NDM, ,Yes
Citrobacter freundii, , ,Yes
ESCHERICHIA COLI, ,Yes,
Klebsiella pneumoniae,KPC, ,Yes
Citrobacter brakii, ,Yes,No
,unk, ,
Klebsiella aerogenes,KPC,Yes,Yes
Candida auris, , ,
Klebsiella pneumonaiae, , ,Yes
Klebsiella oxytoca, ,Yes,No
;
run;Thank you
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
So take a couple of those examples and explain how you got from the input string to the output results.
Why did you pick
Klebsiella aerogenesFrom the first string? Is it because it followed the text
NTE:? If so then why did you remove the 2+ and the other words following NTE:
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Hi Tom,
Klebsiella aerogenes was picked because it's an organism (It falls under the Organism column). It could be anywhere in the string and it doesn't have to do anything with NTE: or 2+. Citrobacter freundii, Citrobacter brakii, and other organisms fall in varied places. I created a list of all organisms (about 22) that should be picked but I'm not sure how I can tackle this.
For the second column, only 5 values can be picked (KPC, NDM, OXA, VIM, and no gene)
The third column picks yes when it says carbapenem resistant dectected or positve and No for carbapenem resistant not detected or No carbapenem resistant detected.
Fourth column same for 3rd but carbapenemase producing instead of carbapenem resistant.
Thank you
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
You are not going to be able to do that with normal coding. The best you could do would be if you had a list of literal strings you considered an ORGANISM. You could then check if that string appears in your character variable.
But there are now language processing tools that might be able to do what you want. Essentially AI or machine learning tools.
https://bmcbioinformatics.biomedcentral.com/articles/10.1186/1471-2105-13-211
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Yes, Tom. That's why I created a list of all possible organisms but I'm not sure how I can use this list to pick them from the string! What code? method?
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Use FINDW() function.
Say your list of organisms is in a dataset named ORGANISMS in a variable named ORGANISM. Then you just need to join your list of NOTES to the ones that match.
proc sql;
create table want as
select a.notes
, b.organism
from have a
left join organisms b
on findw(a.notes,trim(b.organism))
;
quit;
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Thank you Tom. I'll try using this function and see if it works.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Hi Tom,
This worked for the most part. But for #3, #4, and #5 it picked Acinetobacter baumannii complex, Proteus mirabilis, and Morganella morganii instead of Klebsiella pneumoniae. Is there an explanation for that? Below are the data used:
data Organisms;
length Organism $250;
infile datalines dsd truncover;
input Organism $;
datalines4;
Klebsiella pneumoniae
Acinetobacter baumannii complex
Proteus mirabilis
Morganella morganii
Providencia stuartii
Citrobacter freundii
ESCHERICHIA COLI
Klebsiella pneumoniae
Citrobacter brakii
Klebsiella aerogenes
;;;;
run;
data have;
length Notes $256;
infile datalines dsd truncover;
input Notes $;
datalines4;
SPECIMEN NOTES:Collection Method ID: Transport MediaNTE:Heavy Growth Carbapenem Resistant Klebsiella pneumoniae Heavy Growth Multi-Drug Resistant Acinetobacter baumannii complex Moderate Growth ESBL Proteus mirabilis Moderate Growth Morganella morganii Providencia stuartii
SPECIMEN NOTES:Collection Method NTE:75 --99 cfu/ml Citrobacter freundii This CRE isolate demonstrates carbapenemase-producing
SPECIMEN NOTES:119334006 Sputum specimen SPUTUM 1NTE:=4+ CARBAPENEM RESISTANT ESCHERICHIA COLI
NTE:=2+ Klebsiella pneumoniae Carbapenemase-producing Enterobacteriaceae Positivefor Carbapenemase: KPC COLISTIN 2 INTERMEDIATE- Klebsiella pneumoniae to organism column and KPC to Gene-mechanism
SPECIMEN NOTES:Collection Method ID: TMOT - Transport MediaNTE:70 cfu/ml Citrobacter brakii CRE (carbapenem-resistant Enterobacteriaceae) Targeted carbapenemase producing genes NOT detectedOther-Citrobacter brakii to organism, yes to carbapenem-resistant and no to carbapenemase-producing
SPECIMEN NOTES: URC Urine clean catch HL70487 2.5 Urine clean catch
SPECIMEN NOTES:Collection Method ID: TMOT - Transport MediaNTE:Recovered Klebsiella aerogenes ***This CRE isolate demonstrates carbapenemase production.** Positive for NDM gene for carbapenemase production by PCR.Carbapenem resistant organism.
;;;;
run;
proc sql;
create table want as
select a.notes
, b.organism
from have a
left join Organisms b
on findw(a.notes,trim(b.organism))
;
quit;
Thank you
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
You had one of the organisms listed twice in your list.
You might want to ignore the case. You can force everything to uppercase. Also look at the optional arguments for FINDW().
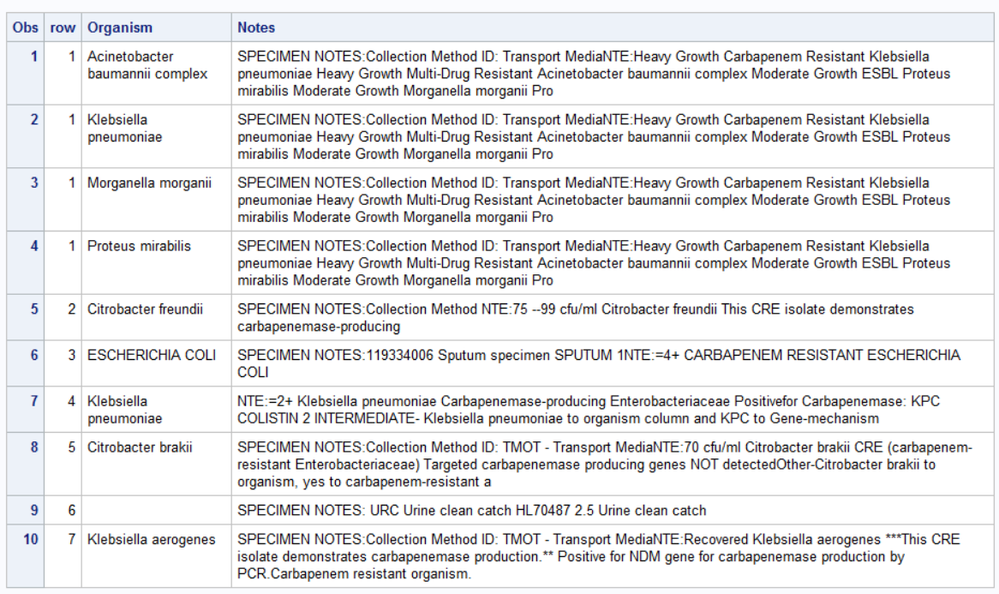
Let's add a row identifier so we can see which notes are which.
data Organisms;
infile datalines truncover;
input Organism $250.;
datalines4;
Klebsiella pneumoniae
Acinetobacter baumannii complex
Proteus mirabilis
Morganella morganii
Providencia stuartii
Citrobacter freundii
ESCHERICHIA COLI
Citrobacter brakii
Klebsiella aerogenes
;;;;
data have;
row+1;
infile datalines truncover;
input Notes $256.;
datalines4;
SPECIMEN NOTES:Collection Method ID: Transport MediaNTE:Heavy Growth Carbapenem Resistant Klebsiella pneumoniae Heavy Growth Multi-Drug Resistant Acinetobacter baumannii complex Moderate Growth ESBL Proteus mirabilis Moderate Growth Morganella morganii Providencia stuartii
SPECIMEN NOTES:Collection Method NTE:75 --99 cfu/ml Citrobacter freundii This CRE isolate demonstrates carbapenemase-producing
SPECIMEN NOTES:119334006 Sputum specimen SPUTUM 1NTE:=4+ CARBAPENEM RESISTANT ESCHERICHIA COLI
NTE:=2+ Klebsiella pneumoniae Carbapenemase-producing Enterobacteriaceae Positivefor Carbapenemase: KPC COLISTIN 2 INTERMEDIATE- Klebsiella pneumoniae to organism column and KPC to Gene-mechanism
SPECIMEN NOTES:Collection Method ID: TMOT - Transport MediaNTE:70 cfu/ml Citrobacter brakii CRE (carbapenem-resistant Enterobacteriaceae) Targeted carbapenemase producing genes NOT detectedOther-Citrobacter brakii to organism, yes to carbapenem-resistant and no to carbapenemase-producing
SPECIMEN NOTES: URC Urine clean catch HL70487 2.5 Urine clean catch
SPECIMEN NOTES:Collection Method ID: TMOT - Transport MediaNTE:Recovered Klebsiella aerogenes ***This CRE isolate demonstrates carbapenemase production.** Positive for NDM gene for carbapenemase production by PCR.Carbapenem resistant organism.
;;;;
proc sql;
create table want as
select a.row
, b.organism
, a.notes
from have a
left join Organisms b
on findw(upcase(a.notes),upcase(trim(b.organism)))
order by row,organism
;
quit;
Learn how use the CAT functions in SAS to join values from multiple variables into a single value.
Find more tutorials on the SAS Users YouTube channel.
SAS Training: Just a Click Away
Ready to level-up your skills? Choose your own adventure.