- Home
- /
- Programming
- /
- Programming
- /
- Re: DO Loop and ARRAY
- RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Hi,
I am trying to create one variable (liverdis)using the values contained in 25 other variables (dx1-dx25). The values are basically ICD 9 codes.
I want to select some specific codes and range of codes of these ICD9s that would form this new variable.
I am not quite sure how I can incorporate the range of these ICD9s in one single array.
This is what I have at present- it contains individual specific codes -07022' '07023' '07044
and the code runs just fine.
data library.CRT1;
set library.CRT1;
array dxmslivedis{25} dx1-dx25;
if cmiss(of dx1-dx25)=25 then call missing(liverdis);
else do;
do i=1 to 25;
if dxmslivedis {i} in('07022' '07023' '07044')
then do;
liverdis=1;
return;
end;
else liverdis=0;
end;
end;
run;
But in the above SAS code, in addition to the specific ICD 9 codes, I also want to add range of codes as this
'4560'<=dxmslivedis<='4562' or '5722'<=dxmslivedis<='5728'
I tried different ways to add but then the above code doesn't run without showing errors.
Any help/suggestions would be welcome!
Thank you!
A
Accepted Solutions
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
I'd use the following. However, be careful when with using less than/greater than when checking for a string variable:
libname library '/folders/myfolders';
data library.crt1;
infile datalines truncover;
input (dx1-dx25) ($);
datalines;
07023 09999
07044
5727
45602
09999
;
data library.CRT1;
set library.CRT1;
array dxmslivedis{25} dx1-dx25;
if cmiss(of dxmslivedis(*))=25 then call missing(liverdis);
else do;
do i=1 to 25;
if (dxmslivedis {i} in ('07022' '07023' '07044'))
or ('4560'<=dxmslivedis(i)<='4562')
or ('5722'<=dxmslivedis(i)<='5728')
then do;
liverdis=1;
return;
end;
else liverdis=0;
end;
end;
run;
Art, CEO, AnalystFinder.com
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
see if this works
data library.CRT1;
set library.CRT1;
array dxmslivedis{25} dx1-dx25;
if cmiss(of dx1-dx25)=25 then call missing(liverdis);
else do;
do i=1 to 25;
if dxmslivedis {i} in('07022' '07023' '07044')then
do;
liverdis=1;
leave;
end;
else liverdis=0;
end;
end;
run;- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Thank you but my question is how to add range of codes as this
'4560'- '4562' and '5722'- 5728'
in addition to the specific ICD 9 codes.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Oh sorry, which variable has those 9 codes. Can you show a sample of your dataset
Also, show a sample of your WANT
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
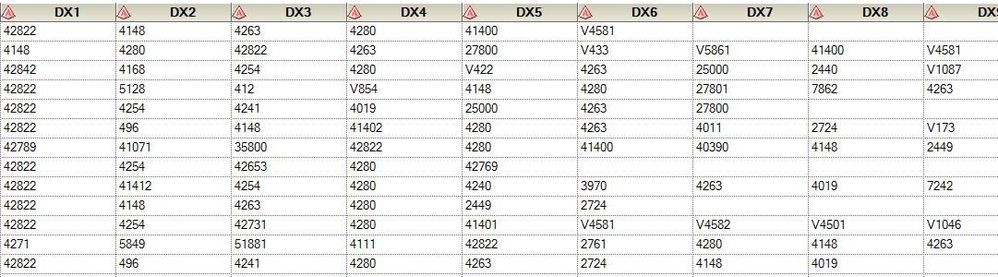
so the same variables used in defining the array : dx1-dx25, these are 25 variables that may contain the icd9 codes included in that range.
The variables look some thing lie below. Hope it makes sense
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
I'd use the following. However, be careful when with using less than/greater than when checking for a string variable:
libname library '/folders/myfolders';
data library.crt1;
infile datalines truncover;
input (dx1-dx25) ($);
datalines;
07023 09999
07044
5727
45602
09999
;
data library.CRT1;
set library.CRT1;
array dxmslivedis{25} dx1-dx25;
if cmiss(of dxmslivedis(*))=25 then call missing(liverdis);
else do;
do i=1 to 25;
if (dxmslivedis {i} in ('07022' '07023' '07044'))
or ('4560'<=dxmslivedis(i)<='4562')
or ('5722'<=dxmslivedis(i)<='5728')
then do;
liverdis=1;
return;
end;
else liverdis=0;
end;
end;
run;
Art, CEO, AnalystFinder.com
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Thank you very much! It worked with no errors. I'd be careful about the string variables.
Appreciate it!
Ashwini
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Requesting you to kindly mark art's answer and close the thread. Thank you!
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
I have a quick question:
In the array above, I want to define range as 25300-25099
('25030'<=dxmslivedis(i)<='25099')
By the very nature of ICD9 code format, the sequence will go like this - 25030,25031,.....,25040,25041,25050....25070...25099. and then the codes for next diagnosis begin from 2501 which I am not interested in.
What will be the best way to define range here if I don't know the upper level code for every diagnosis that wish to code? In this particular case I happen to know the higher/upper level of range will be 25099.
If I use this following sequence, not sure if it will capture all and ONLY the codes between 25030-25099?
('25030'<=dxmslivedis(i)<'2501')
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
@Ashwini_uci wrote:
I have a quick question:
In the array above, I want to define range as 25300-25099
('25030'<=dxmslivedis(i)<='25099')By the very nature of ICD9 code format, the sequence will go like this - 25030,25031,.....,25040,25041,25050....25070...25099. and then the codes for next diagnosis begin from 2501 which I am not interested in.
What will be the best way to define range here if I don't know the upper level code for every diagnosis that wish to code? In this particular case I happen to know the higher/upper level of range will be 25099.
If I use this following sequence, not sure if it will capture all and ONLY the codes between 25030-25099?
('25030'<=dxmslivedis(i)<'2501')
If your comparison is happening in SAS then you can use colon modifier to limit match to shorter string.
('250' <=: dx <=: '253')Or if you are pushing the query into external database then use facts that spaces are less than digits and letter like Z are larger than digits.
('250 ' <= dx <= '253ZZ')Of course this really won't help much with ICD9 codes for Diabetes where the fifth digit is used to distinguish between Type I and Type II diagnosis codes.
if '250' =: dx then do;
diabetes=1;
if substr(dx,5,1) in ('1','3') then type1=1
else if substr(dx,5,1) in ('0','2') then type2=1;
end;
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Also for a range including the following codes
5820,5821..58231..5824, 58281, 58289, is there a way to ask the SAS code to return all ICD9s that start with 582?
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
I'd think that the following satisfies all of your conditions, but you should test some possible exceptions just to be sure:
libname library '/folders/myfolders';
data library.crt1;
infile datalines truncover;
input (dx1-dx25) ($);
datalines;
07023 09999
07044
5727
45602
09999
58231
58289
;
data library.CRT1;
set library.CRT1;
array dxmslivedis{25} dx1-dx25;
if cmiss(of dxmslivedis(*))=25 then call missing(liverdis);
else do;
do i=1 to 25;
if (dxmslivedis {i} in ('07022' '07023' '07044'))
or ('4560'<=dxmslivedis(i)<='4562')
or ('5722'<=dxmslivedis(i)<='5728')
or ('25030'<=dxmslivedis(i)<='25099')
or (dxmslivedis(i)=:'582')
then do;
liverdis=1;
return;
end;
else liverdis=0;
end;
end;
run;
Art, CEO, AnalystFinder.com
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Thank you!
So I want to include these codes below
5820,5821....58231..5824.... 58281, 58289, and there are quite a lot of them.
But is there any way to ask in your SAS code to return all ICD9 numbers that start with 582 and may end up being 4 and 5 digit long numbers, instead of including them all in the code?
or (dxmslivedis(i)=:'582')
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
That was accomplished in the previous code with:
or (dxmslivedis(i)=:'582')
Art, CEO, AnalystFinder.com
Learn how use the CAT functions in SAS to join values from multiple variables into a single value.
Find more tutorials on the SAS Users YouTube channel.
SAS Training: Just a Click Away
Ready to level-up your skills? Choose your own adventure.