- Home
- /
- Programming
- /
- SAS Procedures
- /
- Re: Saving p-values
- RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Hi,
I am trying to do a Monte Carlo simulation of ANCOVA. I generated random variables and simulated data from regressions models below. x is a covariate, g is a grouping variable (0 for a control group, 1 for a treatment group), y is a dependent variable. I am manipulating coefficients. In this syntax, I set the treatment effect on y to be 0, the treatment effect on x to be 0.6, and the effect of x on y to be 1. I also set the number of samples to be 1000 and the sample size to be 800 (about a half being a treatment group).
data MC (drop=i);
call streaminit(12345678);
do sampleID = 1 to 1000;
do i = 1 to 800;
g = rand("Bernoulli", 0.5);
x = 0.6*g + rand("Normal");
y = 1*x + 0*g + rand("Normal");
output;
end;
end;
run;So, after that, I need to test the effect of g on y in each of 1000 samples. I wrote this syntax:
proc glm data = MC;
BY sampleID;
class g;
model y = g;
run;It works but the problem is that I need only p values (for F values) from the output to count how many were less then .05 (which will serve as an estimate of Type I error rate). So, I can't figure out how to put these p-values in a data set, an array, or something like that (extracting p-values by hand for 1000 samples is not an option...).
I would very much appreciate any help and/or feedback. Thank you.
Accepted Solutions
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Here is how to do the counting (for effect g):
proc glm data = MC outstat=myStat noprint;
BY sampleID;
class g;
model y = g x / ss3;
run;
proc sql;
select
sum(prob<0.05) as nb_5pct,
count(prob) as nb_sample,
sum(prob<0.05) / count(prob) as prop_5pct
from myStat
where _SOURCE_ = "g";
quit;
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Start with:
proc glm data = MC outstat=myStat noprint;
BY sampleID;
class g;
model y = g x / ss3;
run;
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Thank you! Now I have one table with all p values!
Last question -- how now to count p that less than 0.05? I though I can use myStat now as a data set and PROB as a variable so that I could simply count through the if statement -- but that doesn't seem to work...
Thank you in advance.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
@Amanda_Lemon wrote:
Thank you! Now I have one table with all p values!
Last question -- how now to count p that less than 0.05? I though I can use myStat now as a data set and PROB as a variable so that I could simply count through the if statement -- but that doesn't seem to work...
Thank you in advance.
What does that code look like, that logic should work fine.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Here is how to do the counting (for effect g):
proc glm data = MC outstat=myStat noprint;
BY sampleID;
class g;
model y = g x / ss3;
run;
proc sql;
select
sum(prob<0.05) as nb_5pct,
count(prob) as nb_sample,
sum(prob<0.05) / count(prob) as prop_5pct
from myStat
where _SOURCE_ = "g";
quit;
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Thank you so much!! It works!
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
An unrelated follow-up question. When I simulate the binary variable g, I use the Bernoulli distribution. I want the two groups to be equal, so I thought that setting the probability to 0.5 will do that. But it produces approximate splits in half (like, 423 and 377). I know I can just assign 0 to g for i from 1 to 400 and 1 to g for i from 401 to 800. But is there a way to _randomly_ assign 0 and 1 to g but have the groups precisely equal (400 and 400)?
Thanks again for your help.
data MC (drop=i);
call streaminit(12345678);
do sampleID = 1 to 1000;
do i = 1 to 800;
g = rand("Bernoulli", 0.5);
x = 0.6*g + rand("Normal");
y = 1*x + 0*g + rand("Normal");
output;
end;
end;
run;- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
There is no advantage in having random g values, as far as I can tell. But if you want, you can do:
data temp (drop=i);
call streaminit(12345678);
do sampleID = 1 to 1000;
do g = 0, 1;
do i = 1 to 400;
rnd = rand("Uniform");
x = 0.6*g + rand("Normal");
y = 1*x + 0*g + rand("Normal");
output;
end;
end;
end;
run;
proc sort data=temp out=mc(drop=rnd);
by sampleId rnd;
run;(untested)
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
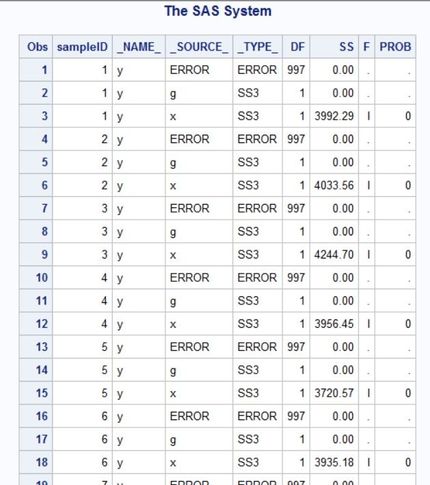
I ran into another problem... Everything works just fine when I simulate a normal distribution but when I changed it to a truncated normal distribution, the PROC GLM statement stopped working... (data are simulated fine). When I print the GLM results, it shows that SS for g were not computed (see the screenshot). Why is that? Thank you in advance.
data MC (keep = x y g sampleID);
Fa = cdf('Normal', -6, 0, 1); /* for a = -6 */
Fb = cdf('Normal', 6, 0, 1); /* for b = 6 */
call streaminit(12345678);
do sampleID = 1 to 1000;
do g = 0, 1;
do i = 1 to 500;
v = Fa + (Fb-Fa)*rand('Uniform'); /* V ~ U(F(a), F(b)) */
x = 0*g + quantile('Normal', v, 0, 1); /* truncated normal on [a,b] */
y = 1*x + 0*g + quantile('Normal', v, 0, 1);
output;
end;
end;
end;
run;
proc glm data = MC outstat=myStat noprint;
BY sampleID;
class g;
model y = g x / ss3;
run;
proc print data = myStat;
run;- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
The problem stems from the fact that y = 2*x in all of your simulated observations. To replicate the structure of your previous simulation you would need:
v = Fa + (Fb-Fa)*rand('Uniform'); /* V ~ U(F(a), F(b)) */
w = Fa + (Fb-Fa)*rand('Uniform'); /* W ~ U(F(a), F(b)) */
x = 0*g + quantile('Normal', v, 0, 1); /* truncated normal on [a,b] */
y = 1*x + 0*g + quantile('Normal', w, 0, 1);
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
April 27 – 30 | Gaylord Texan | Grapevine, Texas
Registration is open
Walk in ready to learn. Walk out ready to deliver. This is the data and AI conference you can't afford to miss.
Register now and save with the early bird rate—just $795!
Learn the difference between classical and Bayesian statistical approaches and see a few PROC examples to perform Bayesian analysis in this video.
Find more tutorials on the SAS Users YouTube channel.
SAS Training: Just a Click Away
Ready to level-up your skills? Choose your own adventure.