- Home
- /
- Solutions
- /
- Data Management
- /
- Re: error when outputting SAS dataset into CSV/excel
- RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
I created a SAS dataset and exported into csv using proc export (code below). When I opened the csv file, there are many rows with invalid, as if the data had shifted somehow. For example, in the year column, I find random text (see picture I also tried the sas export function with the same result. Has anyone ever had this issue? How can I resolve?
PROC EXPORT DATA= DVIS.ABCS15P_PBI /*SAS DATASET TO EXPORT */
OUTFILE= "\\cdc.gov\project\CCID_NCIRD_DBD_RDB\ABCs\Data Vis - BACT FACTs\Phase II\Data-Phase II\ABCS_15P_20170705.csv" /*FILE TO BE CREATED */
DBMS=csv REPLACE;
RUN;
Accepted Solutions
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
It is possible your character varaibles include end of line characters in them that is confusing Excel when reading the CSV file.
For example this line looks strange in your SAS log. Looks like there is a break after the word 'INFO:' . If that break is really in the data then you should probably remove it before trying to send it via a CSV file.
2017,,NM,NM03462,BERNALILLO,0,1,1,1,1,06/07/1928,03/08/2017,88,3,32416,1080,88,1,0,0,0,0,0,1,1 ,2,1,2,2,03/06/2017,03/16/2017,03/16/2017,1,0,0,0,0,0,0,0,0,0,0,0,0,0,,0,,0,0,0,0,0,0,0,0,0,0, 1,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,1,0,0,0,0,0,0,0,0,,,,,,,,,,,,2,2,1,,,,,2,,,,0,0,,,,,0,9,,,,5,9 ,,,0,1,,9,0,0,1,2,,,,,,,0,0,0,,,0,,0,,,,,,,1,0,0,2,6,,,,,2,,,0,0,,0,0,,,,132,13,19.61,,,,,,,,, ,,,,,,,,,LAB03,RM,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,"STILL IN-HOUSE AS OF 3/16 ADDITIONAL CONTACT INFO: JANELLE MONTOYA,",,HP004,87110,N,1,,1,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,, 1,M41824,,,1,,,,,,,,,,,,,,,,,,,,2017007183,3015420898,,,,,03/30/2017,,,,,,,,,1,1,,B,B,,,,,,NT, ,1,B,B,,,,,,,,,,0,,0,0,0,0,0,0,0,,0,,0,0,1,0,0,0,1,0,0,0,0,0,0,0,0,0,0,0,0,0,,0,0,1,0,0,0,0,0, 0,0,0,0,0,0,,0,0,,0,0,0,0,0,0,0,1,,0,1,0,,0,0,0,, 2017,,NM,NM03554,BERNALILLO,0,1,1,1,,06/01/1934,03/25/2017,82,3,30248,1008,82,0,0,0,0,0,0,0,9, ,9,9,,,,03/31/2017,1,1,0,0,0,0,0,0,0,0,0,0,0,0,,0,,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0 ,0,0,0,0,1,0,0,0,0,0,0,0,0,,,,,,,,,,,,,2,0,,,,,,,,1,0,0,,,,,0,,,,,,,,,0,,,,0,0,0,,,,,,,,0,0,0, ,,0,,0,,,,,,,,0,0,,3,,,,,,,,0,0,,0,0,,,,,,0.00,,,,,,,,,,,,,,,,,,LAB03,LB,,,,,,,,,,,,,,,,,,,,,, ,,,,,,,,,,HP006,87114,N,1,,1,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,1,M41825, ,,1,,,,,,,,,,,,,,,,,,,,2017008733,3015420897,,,,,03/30/2017,,,,,,,,,1,1,,C,C,,,,,,C,,1,C,C,,,, ,,,,,,0,,0,0,0,0,0,0,0,,0,,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,,0,0,0,0,0,0,0,0,0,0,0,0,0, 0,,0,0,,0,0,0,0,0,0,0,0,,0,0,0,,0,0,0,,
You could use a step like this to make a copy of the data with the modified content with vertical bars replacing any CR or LF characters in any character variable.
data for_export ;
set DVIS.ABCS15P_PBI ;
array _c _character_;
do over _c ;
_c = translate(_c,'||','0D0A'x);
end;
run;
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Check the text file, open the file in an editor not Excel. Is the file correct or is Excel garbling some how?
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Can you post an example of the beginning of the CSV file? Do not use WORD or EXCEL to open it.
Why not just run a quick SAS data step to dump the lines to the SAS log and copy and paste them into the Insert Code pop-up window.
data _null_;
infile "\\cdc.gov\project\CCID_NCIRD_DBD_RDB\ABCs\Data Vis - BACT FACTs\Phase II\Data-Phase II\ABCS_15P_20170705.csv"
obs=10 ;
input;
put _infile_;
run;That way we can see if the file is ok or if it is Excel of Word that is doing something wrong with it.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
The file is fine, I ran proc freqs in SAS and the output is correct. How do I open the CSV file without using excel?
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Right click the file, Select Open With and choose a text editor, ie NotePad, WordPad, or Notepad++
The workaround for this may be to copy the file and paste it directly into excel and then use the Text to Columns to separate it based on the commas and using the quotes as text qualifiers.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
ok, here's what the file looks like in text:
CAL_YR,STATE,STATE2,STATEID,COUNTY,REASON_DATA,SPECIES,STERILE,STATUS,SEX,BIRTH2,CULT2,AGE,UNI
T,AGEDYS,AGEMTHS,AGEYRS,WHITE,BLACK,AMERIND,ASIAN,PACIFIC,UNKRACE,SUMRACE,RACE,ETHNIC,BESTRACE
,BESTETH,BESTOUT,ADMIT2,DISCHRG2,REPORT2,BLOOD,CSF,PLEURAL,PERINEAL,PERICRD,JOINT,BONE,MUSCLE,
PLACENTA,AMNIOTIC,WOUND,MIDDLEAR,SINUS,OTHSITE,SPECSITE,BODYSITE,SPECBODY,ABORT,ABSCESS,AMNION
,ARTHRIT,BACTEREM,CELL,EMPYEMA,ENDOCARD,EPIGLOT,HUS,MENING,NECRFASC,OSTEOMYE,OTITIS,PERICARD,P
ERITON,PNEU,PEURPER,SEPSHOCK,STSS,AMNISYN,ARTHSYN,BACTSYN,CELLSYN,EMPSYN,ENDOSYN,MENSYN,NEALSY
N,NECFSYN,OSTEOSYN,OTITSYN,PNEUSYN,SESHKSYN,STSSSYN,CARDSYN,HIBDATE5,HIBDATE6,HIBDATE7,HIBDATE
8,MENDATE1,MENDATE2,MENDATE3,MENDATE4,MENDATE5,MENDATE6,TODAYDAT,OUTCOME,INMATCH,SUMSYN_RAW,BA
BYDATE,CIDTDT,SURGDATE,AUDIT,AUTOPSY,BABYDTUNK,BWGHT,CIDT_CULT,CIDTCSF,CIDTOTH,CIDTTYPE,COLLEG
E,DCLOC,DELIVER,ENDOMETR,FLUTST,FOUTCOME,GESTAGE,HEIGHTCM,HEIGHTFT,HEIGHTIN,HIBREC,HIBVACC,HIN
SES,HOSPITAL,HTHPRO,ICU,IHS,MEDICAID,MEDICARE,MENIVACC,MENTYPE1,MENTYPE2,MENTYPE3,MENTYPE4,MEN
TYPE5,MENTYPE6,MILITARY,NOINSUR,NOSEQ,OTHBUG1,OTHBUG2,OTHINSUR,OTHSRC,OTHSYN,PCV13,PCV7,PNEUVA
X,PPV23,PREGES,PREGSTAT,PREVRES,PRISON,PRIVATE,RELAPSE,SEROGROU,SEROTYPE,SOLIRIS,SURGERY,SURGD
TUNK,TRANSFER,TRAUDATE,TRAUDATEUNK,UNHEIGHT,UNKBMI,UNKINFEC,UNKINSUR,UNWEIGHT,VAXNS,VAXREG,WEI
GHTKG,WEIGHTLB,WEIGHTOZ,BMI,CIDT_OTHSITE,CIDT_TYPEOTH,DCFACID,FACID,FLUID,HIBLOT1,HIBLOT2,HIBL
OT3,HIBLOT4,HIBMANU1,HIBMANU2,HIBMANU3,HIBMANU4,HIBNAME1,HIBNAME2,HIBNAME3,HIBNAME4,HOSPID,INI
TIALS,MENLOT1,MENLOT2,MENLOT3,MENLOT4,MENLOT5,MENLOT6,MENMANU1,MENMANU2,MENMANU3,MENMANU4,MENM
ANU5,MENMANU6,MENNAME1,MENNAME2,MENNAME3,MENNAME4,MENNAME5,MENNAME6,OTHHFLU,OTHLOCSP,OTHNEIS,P
REVID,SPECBUG1,SPECBUG2,SPECOTH,SPECSEQ,SPECSYN,SPINSUR,SURVCOM1,SURVCOM2,TRANSID,TXHOSP,ZIP,O
THSEQ,CAL_QTR,CASE_SRC,SUMSYN,NURSHOME,PREGNANT,SPECMALI,SPECORGA,SPECILL2,SPECILL3,MENINAM1,M
ENIDAT1,MENILOT1,MENINAM2,MENIDAT2,MENILOT2,MENINOTH,MENINDES,MENIDAT3,MENILOT3,MENINOT,MENIDA
T4,MENILOT4,PNEUCONJ,PNEUDAT1,PNEUNAM1,PNEUMAN1,PNEULOT1,PNEUDAT2,PNEUNAM2,PNEUMAN2,PNEULOT2,P
NEUDAT3,PNEUNAM3,PNEUMAN3,PNEULOT3,PNEUDAT4,PNEUNAM4,PNEUMAN4,PNEULOT4,TISSUE,INFWEIGHT,BESTGE
ST,DCC,SURGSPEC,SULFA,RIFAMPIN,OXAZONE,OXASCRN,ETEST,PNEUVACC,PNEUNAME,PNEUDATE,UROLOGIC,STER_
ABS,SURGASP,TRACT,DOSENUM,PENSIGN1,OTHRACE,SURV,LABID,EMMTYPEC,EMMTYPE,VIABLE,CLISIGN,CLI,CLIS
IR2,ERYSIGN,ERY,ERYSIR2,TETSIGN,TET,TETSIR2,TYPEC,TYPE,PEN,PENSIR2,TAXSIGN,TAX,TAXSIR2,VANSIGN
,VAN,VANSIR2,ACCESSNO,DASH,ACTSHIP,EXPSHIP,RACTSHIP,REXPSHIP,DATERCVD,STATESEROTYPE,HFLUFINAL,
COM1,COM2,ENTR_DAT,TODAY2,OTHSOURC,EPITYPE,ISOLAVAL,ISOLSHIP,CDCSEROT,GROUP,NMENFINAL,ETTYPE,S
EROSUB,NMENSERO,NCOM1,NCOM2,STATESEROGROUP,EPISERO,SOURCE,CDCSEROG,SEROGRES,PENSIR3,COT,COTSIR
2,TYPE_C,TYPEW,SHIPMENT,SPNT,SPNALL15P,STUDY,AIDS,ALCOHOL,ALCOHOLC,ALCOHOLP,AMPDIGSEQ,AMPLIMBS
EQ,ASCVD,ASPLENIA,ASTHMA,BLUNT,BMT,BURNS,CDIAL,CIRR,CKD,COCHLEAR,COMPDEF,CONTIDX,COPD,CRI,CSD,
CSFLEAK,CVA,DEAF,DEMENTIA,DIABETES,DIALYSIS,GLOBDEFI,HEARSEQ,HEART,HIV,HODGKINS,IVDU,IVDUC,IVD
UP,LEUKEMIA,LUPUS,MUSCLERO,MYELOMA,MYINF,NEPHROT,NEUROMUS,NONE,OBESITY,ODRUGC,ODRUGP,ORGAN,OTH
MALIG,PARASEQ,PARKINS,PENTRAUM,PERNEURO,PLEGIA,PREMIE,PUD,PVD,SCARRSEQ,SEIZSEQ,SEIZURE,SEROTRE
S,SICKLE,SMALIG,SMOKER,SURWOUND,THERAPY,UNKNOWN,UNKSEQ,VARICELL,CFIPS
2017,,CA,EB13530,ALAMEDA,0,1,1,2,2,04/15/1980,02/01/2017,36,3,13441,448,36,0,0,0,0,0,0,0,9,,9,
9,,,,02/07/2017,1,0,0,0,0,0,0,0,0,0,0,0,0,0,,0,,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,1,
0,0,0,0,0,0,0,0,0,0,0,0,,,,,,,,,,,02/08/2017,,2,0,,,,2,,,,1,,,,,,,0,,,,,,,,,,,,,0,0,0,,,,,,,,0
,0,,,,0,,0,,,,,,,,0,0,2,3,,,,,,,,,,0,0,,,,,,,,,,,,,,,,,,,,,,,,,SF057,PD,,,,,,,,,,,,,,,,,,,,,,,
,,,,,,,,,SF057,94544,,1,,1,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,1,M41682,,,
,,,,,,,,,,,,,,,,,,,,VPP4-59,3000736969,,,,,03/16/2017,,,,,,,,,,,,C,C,,,,,,,,,C,C,,,,,,,,,,0,,0
,0,,,0,0,0,,0,,0,0,0,0,0,0,0,,0,0,0,0,0,0,,0,,0,0,0,,0,0,0,0,0,0,0,0,0,0,0,0,0,0,,,0,,0,0,0,0,
0,,,0,,0,0,0,,0,0,,,
2015,,NY,NY17169,ALBANY,0,6,1,1,1,09/25/1928,01/03/2015,86,3,31511,1050,86,1,,,,,,1,1,2,1,2,1,
01/03/2015,01/21/2015,01/06/2015,1,0,0,0,0,0,0,0,0,0,0,0,0,0,,0,,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0
,0,1,0,1,0,0,0,0,0,0,0,0,0,0,0,0,1,1,0,0,,,,,,,,,,,02/20/2015,1,2,2,,,,2,,,,,,,,,,,0,1,,,,,66,
,,,1,,1,0,0,1,,,,,,,,0,0,,,,0,,0,,0,9,0,,3,1,0,1,2,,,,,,2,,,,,0,0,,,,,180,,,,,,,NY14150225,,,,
,,,,,,,,,4,NS,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,4,12211,,1,SURV15,2,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,
,,,,,,,,,,,,,,,,,,,,,,,,,,,1,20152208,,,1,,,,,0.03,S,,,,,003,0.03,,,,,,,,,,,,,,,,,,,,,,,1,,,,,
,,,,,,,,,,S,0.12,S,,003,,1,1,,0,,0,0,,,1,0,0,,0,,0,0,1,0,0,0,0,,0,0,0,0,0,1,,0,,0,0,0,,0,0,0,0
,0,0,0,0,0,0,0,0,0,0,,,0,,0,0,0,0,0,,,0,,0,0,0,,0,0,,,
2017,,NM,NM03462,BERNALILLO,0,1,1,1,1,06/07/1928,03/08/2017,88,3,32416,1080,88,1,0,0,0,0,0,1,1
,2,1,2,2,03/06/2017,03/16/2017,03/16/2017,1,0,0,0,0,0,0,0,0,0,0,0,0,0,,0,,0,0,0,0,0,0,0,0,0,0,
1,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,1,0,0,0,0,0,0,0,0,,,,,,,,,,,,2,2,1,,,,,2,,,,0,0,,,,,0,9,,,,5,9
,,,0,1,,9,0,0,1,2,,,,,,,0,0,0,,,0,,0,,,,,,,1,0,0,2,6,,,,,2,,,0,0,,0,0,,,,132,13,19.61,,,,,,,,,
,,,,,,,,,LAB03,RM,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,"STILL IN-HOUSE AS OF 3/16
ADDITIONAL CONTACT INFO:
JANELLE MONTOYA,",,HP004,87110,N,1,,1,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,
1,M41824,,,1,,,,,,,,,,,,,,,,,,,,2017007183,3015420898,,,,,03/30/2017,,,,,,,,,1,1,,B,B,,,,,,NT,
,1,B,B,,,,,,,,,,0,,0,0,0,0,0,0,0,,0,,0,0,1,0,0,0,1,0,0,0,0,0,0,0,0,0,0,0,0,0,,0,0,1,0,0,0,0,0,
0,0,0,0,0,0,,0,0,,0,0,0,0,0,0,0,1,,0,1,0,,0,0,0,,
2017,,NM,NM03554,BERNALILLO,0,1,1,1,,06/01/1934,03/25/2017,82,3,30248,1008,82,0,0,0,0,0,0,0,9,
,9,9,,,,03/31/2017,1,1,0,0,0,0,0,0,0,0,0,0,0,0,,0,,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0
,0,0,0,0,1,0,0,0,0,0,0,0,0,,,,,,,,,,,,,2,0,,,,,,,,1,0,0,,,,,0,,,,,,,,,0,,,,0,0,0,,,,,,,,0,0,0,
,,0,,0,,,,,,,,0,0,,3,,,,,,,,0,0,,0,0,,,,,,0.00,,,,,,,,,,,,,,,,,,LAB03,LB,,,,,,,,,,,,,,,,,,,,,,
,,,,,,,,,,HP006,87114,N,1,,1,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,1,M41825,
,,1,,,,,,,,,,,,,,,,,,,,2017008733,3015420897,,,,,03/30/2017,,,,,,,,,1,1,,C,C,,,,,,C,,1,C,C,,,,
,,,,,,0,,0,0,0,0,0,0,0,,0,,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,,0,0,0,0,0,0,0,0,0,0,0,0,0,
0,,0,0,,0,0,0,0,0,0,0,0,,0,0,0,,0,0,0,,
2015,,GADHR,GA09723,BURKE,0,1,1,4,2,03/25/2013,12/30/2015,2,3,1010,33,2,1,,,,,,1,1,2,1,2,1,12/
30/2015,01/04/2016,04/01/2016,1,,,,,,,,,,,,,,,,,,,,,1,,,,,,,,,,,,,,0,,0,0,1,0,0,0,0,0,0,0,0,0,
0,0,0,,,,,,,,,,,06/27/2017,1,2,1,,,,2,,,,,,,,,,,,2,,,,,,,,,1,,2,,,0,2,0,0,0,0,0,0,,,0,,,,,,,,,
,,3,,0,,,5,,,,,1,,,1,,,,1,,,,,,,,,,,,,,,,,,,,,,,,GA237,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,GA237,GA
158,,N,4,SURV15,1,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,1,M39181,,,,,,,,,,,,
,,,,,,,,,,,16C0140259,3000505554,,,,,03/02/2016,,,,,,,,,,,,W,W,,,,,,,,,W135,W135,,,,,,,,,,0,,0
,0,0,0,0,0,0,,0,,0,0,0,0,0,,0,,0,0,0,0,0,0,,0,0,0,0,0,,0,0,0,0,0,0,,0,,1,0,0,0,0,,0,0,,0,0,0,,
,0,0,0,,0,0,0,,0,,0,,
2015,,GAMSA,GA76522,CARROLL,0,6,1,1,2,12/17/1956,01/05/2015,58,3,21203,706,58,1,0,0,0,0,0,1,1,
2,1,2,2,01/05/2015,01/06/2015,01/08/2015,1,0,0,0,0,0,0,0,0,0,0,0,0,0,,0,,0,0,0,0,0,0,0,0,0,0,0
,0,0,0,0,0,1,0,0,0,0,0,0,0,0,0,0,0,0,0,0,1,0,0,0,,,,,,,,,,,,2,2,1,,,,2,2,,,,,,,,,,0,2,,,,,65,,
,,1,,1,0,0,0,,,,,,,,0,0,,,,0,,0,0,0,9,0,,3,1,0,1,2,,,,,,2,,,0,0,0,0,0,0,,70,,,,,,,,,,,,,,,,,,,
,,GA045,SVH,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,GA052,30180,,1,SURV15,1,,,,,,,,,,,,,,,,,,,,,,,,,,,,
,,,,,,,,,,,,,,,,,,,,,,,,,,,,,1,20151632,,,1,,,,,32,R,,,,,003,0.06,,,,,,,,,,,,,,,,,,,,,,,1,,,,,
,,,,,,,,,,S,0.12,S,,003,,1,1,,0,,1,0,,,0,0,0,,0,,0,0,0,0,0,,0,0,0,0,0,0,0,0,0,0,,0,0,0,,0,0,0,
0,0,0,,0,0,0,0,0,0,0,,,0,,0,0,0,,,,,0,,0,0,1,,0,0,,,
2015,,GAMSA,GA76555,CARROLL,0,6,1,1,2,11/09/1940,01/11/2015,74,3,27091,903,74,1,0,0,0,0,0,1,1,
2,1,2,1,01/11/2015,01/15/2015,01/15/2015,1,0,0,0,0,0,0,0,0,0,0,0,0,0,,0,,0,0,0,0,0,0,0,0,0,0,0
,0,0,0,0,0,1,0,0,0,0,0,0,0,0,0,0,0,0,0,0,1,0,0,0,,,,,,,,,,,,1,2,1,,,,2,,,,,,,,,,,0,2,,,120,,,,
,,1,,2,0,0,1,,,,,,,,0,0,,22,,0,,0,0,0,1,0,,3,1,0,1,2,,,,,,2,,,0,0,0,0,0,1,,64,,,,,,,,,,,,,,,,,
,,,,GA045,SVH,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,GA045,30179,,1,SURV15,1,,,,,,,,,,,,,,,,,,,,,,,,,,
,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,1,20151886,,,1,,,,,8,R,,,,,17F,0.03,,,,,,,,,,,,,,,,,,,,,,,1,,,,
,,,,,,,,,,,S,0.25,S,,17F,,1,1,,0,,0,0,,,0,0,0,,0,,0,0,0,0,0,,0,0,0,0,0,0,0,0,0,0,,0,0,0,,0,0,0
,0,0,0,,0,0,0,0,0,0,0,,,0,,0,0,0,,,,,0,,0,0,0,,0,0,,,
NOTE: 10 records were read from the infile "\\cdc.gov\project\CCID_NCIRD_DBD_RDB\ABCs\Data
Vis - BACT FACTs\Phase II\Data-Phase II\ABCS15P_20170707N.csv".
The minimum record length was 25.
The maximum record length was 3265.
NOTE: DATA statement used (Total process time):
real time 0.06 seconds
cpu time 0.04 seconds- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
It is possible your character varaibles include end of line characters in them that is confusing Excel when reading the CSV file.
For example this line looks strange in your SAS log. Looks like there is a break after the word 'INFO:' . If that break is really in the data then you should probably remove it before trying to send it via a CSV file.
2017,,NM,NM03462,BERNALILLO,0,1,1,1,1,06/07/1928,03/08/2017,88,3,32416,1080,88,1,0,0,0,0,0,1,1 ,2,1,2,2,03/06/2017,03/16/2017,03/16/2017,1,0,0,0,0,0,0,0,0,0,0,0,0,0,,0,,0,0,0,0,0,0,0,0,0,0, 1,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,1,0,0,0,0,0,0,0,0,,,,,,,,,,,,2,2,1,,,,,2,,,,0,0,,,,,0,9,,,,5,9 ,,,0,1,,9,0,0,1,2,,,,,,,0,0,0,,,0,,0,,,,,,,1,0,0,2,6,,,,,2,,,0,0,,0,0,,,,132,13,19.61,,,,,,,,, ,,,,,,,,,LAB03,RM,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,"STILL IN-HOUSE AS OF 3/16 ADDITIONAL CONTACT INFO: JANELLE MONTOYA,",,HP004,87110,N,1,,1,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,, 1,M41824,,,1,,,,,,,,,,,,,,,,,,,,2017007183,3015420898,,,,,03/30/2017,,,,,,,,,1,1,,B,B,,,,,,NT, ,1,B,B,,,,,,,,,,0,,0,0,0,0,0,0,0,,0,,0,0,1,0,0,0,1,0,0,0,0,0,0,0,0,0,0,0,0,0,,0,0,1,0,0,0,0,0, 0,0,0,0,0,0,,0,0,,0,0,0,0,0,0,0,1,,0,1,0,,0,0,0,, 2017,,NM,NM03554,BERNALILLO,0,1,1,1,,06/01/1934,03/25/2017,82,3,30248,1008,82,0,0,0,0,0,0,0,9, ,9,9,,,,03/31/2017,1,1,0,0,0,0,0,0,0,0,0,0,0,0,,0,,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0 ,0,0,0,0,1,0,0,0,0,0,0,0,0,,,,,,,,,,,,,2,0,,,,,,,,1,0,0,,,,,0,,,,,,,,,0,,,,0,0,0,,,,,,,,0,0,0, ,,0,,0,,,,,,,,0,0,,3,,,,,,,,0,0,,0,0,,,,,,0.00,,,,,,,,,,,,,,,,,,LAB03,LB,,,,,,,,,,,,,,,,,,,,,, ,,,,,,,,,,HP006,87114,N,1,,1,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,1,M41825, ,,1,,,,,,,,,,,,,,,,,,,,2017008733,3015420897,,,,,03/30/2017,,,,,,,,,1,1,,C,C,,,,,,C,,1,C,C,,,, ,,,,,,0,,0,0,0,0,0,0,0,,0,,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,,0,0,0,0,0,0,0,0,0,0,0,0,0, 0,,0,0,,0,0,0,0,0,0,0,0,,0,0,0,,0,0,0,,
You could use a step like this to make a copy of the data with the modified content with vertical bars replacing any CR or LF characters in any character variable.
data for_export ;
set DVIS.ABCS15P_PBI ;
array _c _character_;
do over _c ;
_c = translate(_c,'||','0D0A'x);
end;
run;
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
I would never let Excel open a CSV file automatically. Perhaps if I knew that it only contain numbers and did not have any dates or character variables.
Start Excel and then tell it to open the file. In Excel 2010 it is under the DATA menu.
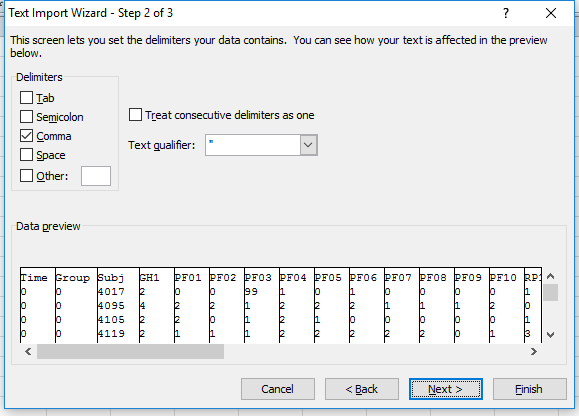
Once you pick a file and tell it that the file is delimited it will prompt you for what your delimiter is.
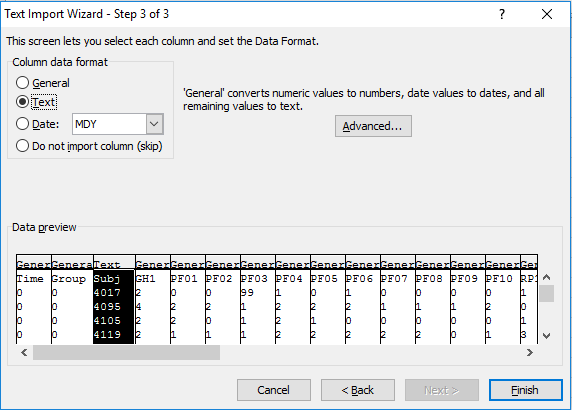
You can then tell it how to treat each column. So if you have a text field that might have values that look like numbers you can tell Excel to treat the whole column as character.
Need to connect to databases in SAS Viya? SAS’ David Ghan shows you two methods – via SAS/ACCESS LIBNAME and SAS Data Connector SASLIBS – in this video.
Find more tutorials on the SAS Users YouTube channel.