- Home
- /
- Solutions
- /
- Data Management
- /
- Re: Conditional coding
- RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Hello,
I have a dataset about cancer. SEER has some conditions in defining brain tumors as benign.
For other tumors such as breast, cervix, and other cancers I used:
if Seer_site_group in ( 26000 ) and beh=2
then CASite67= 1901 ; *Breast, In Situ;
if Seer_site_group in ( 26000 ) and beh=3
then CASite67= 1902 ; *Breast, Malignant;
if Seer_site_group in ( 27010 )
then CASite67= 2000 ; *Cervix;
if Seer_site_group in ( 27020)
then CASite67= 2101 ; *Corpus Uteri;
if Seer_site_group in ( 27030 )
then CASite67= 2102 ; *Uteri, NOS;
if Seer_site_group in ( 27040 )
then CASite67= 2200 ; *Ovary;
For brain cancer I used:
if Seer_site_group in ( 31010 ) and beh=2
then CASite67= 3101 ; *Brain, Benign;
if p in ( 31010 ) and beh=3
then CASite67= 3102 ; *Brain, Malignant;
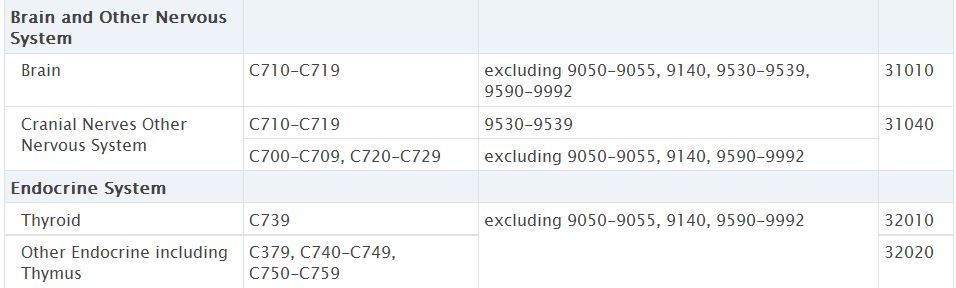
Then I realized that brain tumor in the "Seer_site_group" with code (32020) captures more than just C751-C753. So the code should involve something like that:
IF "beh" = 0 or 1; AND Primary_site = C751, C752, C753; AND histology_icdo3 NOT = 9050-9055, 9140, 9590-9992; THEN = "Benign Brain"
but I'm not sure how to integrate that in one SAS doable statement along with the other cancer statements.
Thank you.
Accepted Solutions
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
@mayasak wrote:
Thank you @FreelanceReinh, please note that histology_icdo3 is a character value too with a length of exactly 4 for all of them.
This can be dealt with easily:
For example, if you know for sure that this character variable always ("always" in the relevant observations, not necessarily in general!) contains 4 digits (from '0' to '9'), you could replace the corresponding condition by
input(histology_icdo3, 4.) not in (9050:9055 9140 9590:9992)
If, however, there is a possibility that some of the codes are alphanumeric and, e.g., '959A' is regarded as belonging to the range "9590-9992," you could use inequalities with regard to alphabetical order:
not ('9050'<=histology_icdo3<='9055' | histology_icdo3='9140' | '9590'<=histology_icdo3<='9992')
This is equivalent to ballardw's format approach.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Sorry I forgot that I had this statement too regarding the question.
if Seer_site_group in ( 32020 )
then CASite67= 3400 ; *Other Endocrine including Thymus;
Thanks
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Hello @mayasak,
Syntactically correct would be this:
if beh in (0 1) and primary_site in ('C751' 'C752' 'C753')
and histology_icdo3 not in (9050:9055 9140 9590:9992) then CASite67 = 3101; *Brain, Benign;(assuming that primary_site is a character variable with values such as those listed and histology_icdo3 is numeric, so that the abbreviations for ranges of integers like 1:5 for 1 2 3 4 5 are valid).
Please adapt and combine this with a criterion about Seer_site_group if necessary and appropriate. It seems confusing to me that you write 'brain tumor in the "Seer_site_group" with code (32020)' while code 32020 is rather subordinate to the endocrine system.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Thank you FreelanceReinhard, please note that histology_icdo3 is a character value too with a length of exactly 4 for all of them.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
@mayasak wrote:
Thank you @FreelanceReinh, please note that histology_icdo3 is a character value too with a length of exactly 4 for all of them.
This can be dealt with easily:
For example, if you know for sure that this character variable always ("always" in the relevant observations, not necessarily in general!) contains 4 digits (from '0' to '9'), you could replace the corresponding condition by
input(histology_icdo3, 4.) not in (9050:9055 9140 9590:9992)
If, however, there is a possibility that some of the codes are alphanumeric and, e.g., '959A' is regarded as belonging to the range "9590-9992," you could use inequalities with regard to alphabetical order:
not ('9050'<=histology_icdo3<='9055' | histology_icdo3='9140' | '9590'<=histology_icdo3<='9992')
This is equivalent to ballardw's format approach.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
I ran this as you've stated:
if Seer_site_group in ( 31010 32020 ) and beh in (0 1) and primary_site in ('C751' 'C752' 'C753') and input(histology_icdo3, 4.) not in (9050:9055 9140 9590:9992)
then CASite67= 3101 ; *Brain, Benign;
if Seer_site_group in ( 31010 ) and beh=3
then CASite67= 3102 ; *Brain, Malignant;
there was no error in the log however, when I ran proc freq for casite67, 3101 was missing in the table.
2900 97
3000 338
3102 1109
3200 2477
I also ran the proc freq with:
if Seer_site_group in ( 31010 32020 ) and beh in (0 1) and primary_site in ('C751' 'C752' 'C753')
then CASite67= 3101 ; *Brain, Benign;
and had the same problem.
It worked only with:
if Seer_site_group in ( 31010 32020 ) and beh in (0 1)
then CASite67= 3101 ; *Brain, Benign;
2900 97
3000 338
3101 285
3102 1109
3200 2477
Thank you
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
If you have a condition that involves multiple comparisons then you may be looking at using the IN operator.
Basically you provide the list of values inside paratheses:
If X in (23 24 25 26) then (do something)
or for character comparisons:
If y in ('23' '24' '25' '26') then ...
You can use NOT as well
If y not in ('23' '24' '25' '26') then ...
If you want a sequence of values that are numeric (actual numbers not character values that look like numbers) you can use
if X in (3:6 9 15) to match values of 3 to 6 and 9 and 15.
So IF "beh" = 0 or 1; AND Primary_site = C751, C752, C753"
would become
If beh in (0,1) and Primary_site in ('C751' 'C752' 'C753') ...
I suspect you 9050-9055, 9140, 9590-9992 values are likely to be character values and even writing an IN statement gets long and the range indicator : is not allowed for character comparisons.
You may be able to use a custom format. Please look at the following code, run it and examine the log.
proc format library=work;
value $code
'9050'-'9055', '9140', '9590'-'9992' = 'Match'
other ='No match';
run;
data _null_;
length x $ 8.;
do x= '9050','9051','9166','9593','95936','960';
if put(x,$code.) = 'Match' then put "Found" x=;
end;
run;Note that you test a character value. But character value ranges basically will accept anything that matches the first 4 characters if that is all that are used in the defining Format. So 95936 is considered to be between 9590 and 9992 because the 95936 part is. Also the 3 character string matches the first 3 that appear in that range.
So depending on your actual range of values of your histology variable (basically all of them exactly 4 characters) you may be able to include a condition of
and put(histology_icd03,$code.)='No match' with the previous.
Or make a very long IN clause.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Thank you ballardw. The histology is a character variable and it has exactly 4 characters for all.
I ran the
proc format library=work; value $code '9050'-'9055', '9140', '9590'-'9992' = 'Match' other ='No match'; run; data _null_;length x $ 8.; do x= '9050','9051','9166','9593','95936','960'; if put(x,$code.) = 'Match' then put "Found" x=; end; run;as it is after I read the data as follows:
libname temp "I:\cancer";
data CA_test;
set cancer_data;
run;
and then I added
if Seer_site_group in (32020) and beh in (0,1) and Primary_site in ('C751' 'C752' 'C753') and (histology_icd03,$code.)='No match';
then CASite67= 3101 ; *Brain, Benign;
I have the following error:
if Seer_site_group in ( 32020 ) and beh in (0,1) and Primary_site in ('C751' 'C752' 'C753') and
2688! (histology_icd03 $code)='No match'
-
388
200
ERROR 388-185: Expecting an arithmetic operator.
ERROR 200-322: The symbol is not recognized and will be ignored.
I'm not sure what is done with the format statement and what has gone wrong. Do I have to run as it is or customize it somewhere according to my dataset.
Thank you
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
You forgot to type the name of the PUT function:
put(histology_icd03, $code.)
Edit: ... among other things.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Your error:
if Seer_site_group in ( 32020 ) and beh in (0,1) and Primary_site in ('C751' 'C752' 'C753') and
(histology_icd03 $code)='No match'Does not match the posted code:
if Seer_site_group in (32020) and beh in (0,1) and Primary_site in ('C751' 'C752' 'C753') and (histology_icd03,$code.)='No match';
Posted code
Missing Put(histology_ (etc.)) AND includes a ; before the then.
The posted error is missing Put, comma separating variable and format, and period at end of format .
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
I rerun it with the correction. It went through with no errors but as I mentioned to FreelanceReinhard in my last reply that 3101 was missing when I ran the proc freq for casite67:
2900 97
3000 338
3102 1109
3200 2477
3300 4882
It worked when only beh condition added.
Thank you a lot
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
We don't have your data. So, you'll have to check why the two IF conditions for casite67=3101 are never met. For example, you can apply the weak criterion that produces observations with casite67=3101, let's call the resulting dataset RESULT, and then apply PROC FREQ to variables primary_site and histology_icdo3. Example:
proc freq data=result;
where casite67=3101;
format _all_;
tables primary_site;
run;- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Freq results from:
proc freq data=result;
where casite67=3101;
format _all_;
tables beh primary_site histology_icdo3 /nocol nocum nopercent norow;
run;
Beh Frequency
0 197
1 215
primary_site
primary_site Frequency
C710 19
C711 38
C712 36
C713 21
C714 16
C715 35
C716 73
C717 32
C718 9
C719 133
histology_icdo3
histology_icdo3 Frequency
8000 132
8001 1
8680 1
8824 1
8850 2
9084 3
9120 31
9121 42
9122 1
9150 4
9161 53
9350 4
9351 4
9361 1
9383 20
9384 3
9390 18
9394 3
9400 2
9413 4
9440 2
9444 2
9490 1
9493 2
9505 23
9506 4
9540 5
9560 43
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
@mayasak wrote:
(...)
primary_siteprimary_site Frequency
C710 19
C711 38
C712 36
C713 21
C714 16
C715 35
C716 73
C717 32
C718 9
C719 133
histology_icdo3(...)
But you are no longer wondering why a criterion stronger than
primary_site in ('C751' 'C752' 'C753')
resulted in zero observations, are you?
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Sure I'm not ![]()
Thank you
April 27 – 30 | Gaylord Texan | Grapevine, Texas
Registration is open
Walk in ready to learn. Walk out ready to deliver. This is the data and AI conference you can't afford to miss.
Register now and save with the early bird rate—just $795!
Need to connect to databases in SAS Viya? SAS’ David Ghan shows you two methods – via SAS/ACCESS LIBNAME and SAS Data Connector SASLIBS – in this video.
Find more tutorials on the SAS Users YouTube channel.



