- Home
- /
- Programming
- /
- Graphics
- /
- proc gchart: adding 'subgroup' gives different error bars
- RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Dear all,
When adding 'subgroup' to the gchart options, in order to allow bars have pre-defined patterns, the error bars are suddenly wrongly calculated. I do not know what the reason would be, and how to solve the problem for this simple graph...
All help more than welcome
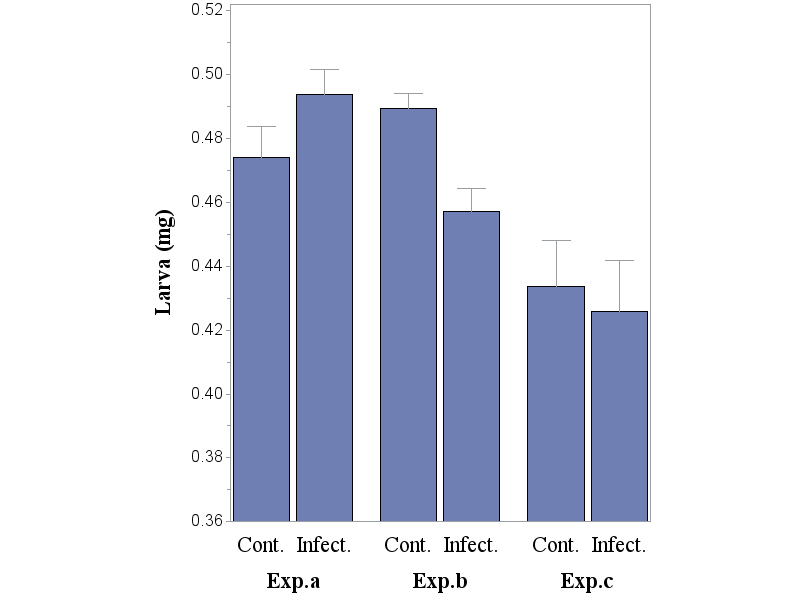
(A) below the code and barchart without subgroup, and correctly calculated error bars:
goptions reset=all;
proc gchart data = cornell.transweightfinalchart;
legend label=(h=1.5) VALUE=(h=1.5);
axis1 label=(f='Times new roman/bo' h=2 a=90 "Larva (mg)" ) order=(0.36 to 0.52 by 0.02) value=(h=1.5) minor=(n=1);
axis2 label=none value=(h=2 f='Times new roman');
axis3 label=none value=(h=2 f='Times new roman/bo');
vbar Treatment/
group=Experiment
sumvar=L
errorbar=top
clm=68.26
width= 8
space= 1
gspace= 3
legend= legend
type=mean
raxis= axis1
maxis=axis2
gaxis=axis3;
where treatment ne '';
run;
quit;
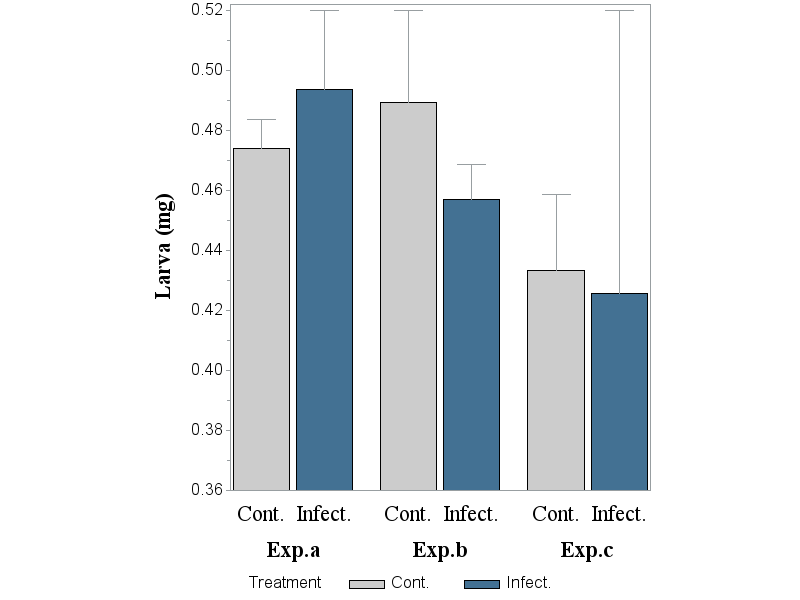
(B) Below a subgroup, named 'Treatment' is added in order to adjust the pattern of the bar that represents either the the Cont. or the Infect. group (i.e. also pattern1 and pattern2 has been added). Error bars: not correct
proc gchart data = cornell.transweightfinalchart;
pattern1 v=solid color=graycc;
pattern2 v=solid color=cx437193;
legend label=(h=1.5) VALUE=(h=1.5);
axis1 label=(f='Times new roman/bo' h=2 a=90 "Larva (mg)" ) order=(0.36 to 0.52 by 0.02) value=(h=1.5) minor=(n=1);
axis2 label=none value=(h=2 f='Times new roman');
axis3 label=none value=(h=2 f='Times new roman/bo');
vbar Treatment/
group=Experiment
sumvar=L
errorbar=top
clm=68.26
subgroup=Treatment
width= 8
space= 1
gspace= 3
legend= legend
type=mean
raxis= axis1
maxis=axis2
gaxis=axis3;
where treatment ne '';
run;
quit;
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
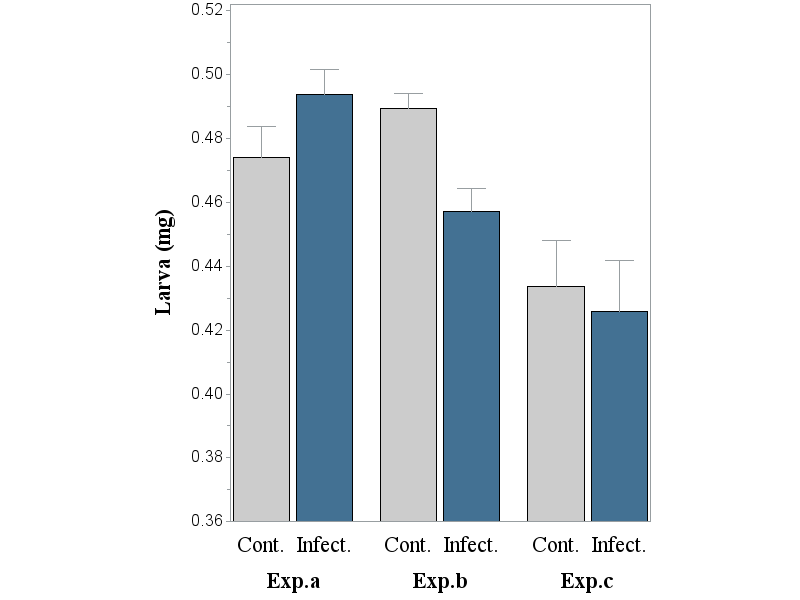
Take away the SUBGROUP and add PATTERNID=MIDPOINT to see if you get what you expect.
Hope this helps!
Dan
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Wow Dan,
this is THE solution, indeed! I do not know why, but it worked.
Thank you so much!
Dieter
Learn how use the CAT functions in SAS to join values from multiple variables into a single value.
Find more tutorials on the SAS Users YouTube channel.
SAS Training: Just a Click Away
Ready to level-up your skills? Choose your own adventure.