- Home
- /
- Programming
- /
- Graphics
- /
- Re: Why the cell border is still there?
- RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Hi there.
I am defining a template for the lattice layout of my heatmaps with the polygonplot statement. I am 95% satisfied with the output. However, the cell border is the problem I cannot resolve. I simply want to remove the cell borders. I used "border=false" in each cell. I also tried to use "borderattrs=(color=white) backgroundcolor=white" to blend the border into background. But neither works. Below is the program I used.
proc template;
define statgraph polygons_all;
begingraph /designwidth=2000px border=false;
entrytitle textattrs=(size=12pt color=black weight=bold) 'Need Gaps';
entryfootnote textattrs=(size=12pt color=black)
"Need gap = Differences between the demand and supply in ZCTAs";
legenditem type=marker name="R" / label="Not rural areas"
lineattrs=(color=gray)
markerattrs=(symbol=squarefilled color=gray size=10pt);
legenditem type=marker name="U" / label="Not urban areas"
lineattrs=(color=gray)
markerattrs=(symbol=squarefilled color=lightgray size=10pt);
discreteattrmap name="b_R" / ignorecase=true;
value "Need gap, [0.31, 1.57]"/
fillattrs=GraphData1(color=A581A22FF);
value "Need gap, [0.12, 0.31)"/
fillattrs=GraphData2(color=A985159FF);
value "Need gap, [0.05, 0.12)"/
fillattrs=GraphData3(color=AB27C83FF);
value "Need gap, [0, 0.05)"/
fillattrs=GraphData4(color=AD8BEC1FF);
value "Need gap, (-0.47, 0)"/
fillattrs=GraphData5(color=A80CDC433);
value "Need gap, (-0.99, -0.47]"/
fillattrs=GraphData6(color=A26A69A33);
value "Need gap, (-2.71, -0.99]"/
fillattrs=GraphData7(color=A00897B33);
value "Need gap, [-8.88, -2.71]"/
fillattrs=GraphData8(color=A004D4033);
value "Not rural areas"/
fillattrs=GraphData9(color=Gray);
value "Missing"/
fillattrs=GraphData10(color=WHITE);
enddiscreteattrmap;
discreteattrvar attrvar=lvlR var=lvlR attrmap="b_R";
discreteattrmap name="b_U" / ignorecase=true;
value "Need gap, [0.31, 1.57]"/
fillattrs=GraphData1(color=A581A22FF);
value "Need gap, [0.12, 0.31)"/
fillattrs=GraphData2(color=A985159FF);
value "Need gap, [0.05, 0.12)"/
fillattrs=GraphData3(color=AB27C83FF);
value "Need gap, [0, 0.05)"/
fillattrs=GraphData4(color=AD8BEC1FF);
value "Need gap, (-0.47, 0)"/
fillattrs=GraphData5(color=A80CDC433);
value "Need gap, (-0.99, -0.47]"/
fillattrs=GraphData6(color=A26A69A33);
value "Need gap, (-2.71, -0.99]"/
fillattrs=GraphData7(color=A00897B33);
value "Need gap, [-8.88, -2.71]"/
fillattrs=GraphData8(color=A004D4033);
value "Not urban areas"/
fillattrs=GraphData11(color=lightGray);
value "Missing"/
fillattrs=GraphData10(color=WHITE);
enddiscreteattrmap;
discreteattrvar attrvar=lvlU var=lvlU attrmap="b_U";
discreteattrmap name="b_all" / ignorecase=true;
value "Need gap, [0.31, 1.57]"/
fillattrs=GraphData1(color=A581A22FF);
value "Need gap, [0.12, 0.31)"/
fillattrs=GraphData2(color=A985159FF);
value "Need gap, [0.05, 0.12)"/
fillattrs=GraphData3(color=AB27C83FF);
value "Need gap, [0, 0.05)"/
fillattrs=GraphData4(color=AD8BEC1FF);
value "Need gap, (-0.47, 0)"/
fillattrs=GraphData5(color=A80CDC433);
value "Need gap, (-0.99, -0.47]"/
fillattrs=GraphData6(color=A26A69A33);
value "Need gap, (-2.71, -0.99]"/
fillattrs=GraphData7(color=A00897B33);
value "Need gap, [-8.88, -2.71]"/
fillattrs=GraphData8(color=A004D4033);
value "Missing"/
fillattrs=GraphData10(color=WHITE);
enddiscreteattrmap;
discreteattrvar attrvar=lvlA var=lvlA attrmap="b_all";
layout lattice/columns=4 rows=1 COLUMNWEIGHTS=(.28 .28 .28 .16)
border=false backgroundcolor=white opaque=true;
cell;
cellheader;
entry "Rural areas"/textattrs=(size=12pt color=black);
endcellheader;
layout overlay/xaxisopts=(display=none) yaxisopts=(display=none)
border=false backgroundcolor=white opaque=true;
polygonplot X=x Y=y ID=ID/
group=lvlR display=(fill outline) dataskin=pressed
name="g_R"
OUTLINEATTRS=(color=white);
endlayout;
endcell;
/* The second cell for the map of "Urban areas". */
cell;
cellheader;
entry "Urban areas"/textattrs=(size=12pt color=black);
endcellheader;
layout overlay/xaxisopts=(display=none) yaxisopts=(display=none)
border=false borderattrs=(color=white) backgroundcolor=white opaque=true;
polygonplot X=x Y=y ID=ID/
group=lvlU display=(fill outline) dataskin=pressed
name="g_U"
OUTLINEATTRS=(color=white)
;
endlayout;
endcell;
cell;
cellheader;
entry "All areas"/textattrs=(size=12pt color=black);
endcellheader;
layout overlay/xaxisopts=(display=none) yaxisopts=(display=none)
border=false backgroundcolor=white opaque=true;
polygonplot X=x Y=y ID=ID/
group=lvlA display=(fill outline) dataskin=pressed
name="g_all"
OUTLINEATTRS=(color=white);
endlayout;
endcell;
discretelegend "g_all" "R" "U"/ LOCATION=outside halign=right
border=false
title="Gap categories" titleattrs=(color=black size=12pt)
across = 1 valueattrs=(color=black size=12pt)
autoitemsize=true;
endlayout;
endgraph;
end;
run;
proc sgrender data=gap1_map template=polygons_all;
format lvlR flvlR. lvlU flvlU. lvlA flvlA. ;
run;
Accepted Solutions
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
If you are talking about the borders around each cell in the lattice then you are probably looking to turn off the WALL, not the border. In the layout overlay options specify WALLDISPLAY=none (or off?). The wall is normally drawn around each overlay layout space and covers the x/y/x2/y2 axes.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Hello,
May you please send me a screenshot of your output so I can see the issue that you are having.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
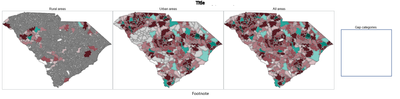
Thanks for your reply.
Here is the final output.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
If you are talking about the borders around each cell in the lattice then you are probably looking to turn off the WALL, not the border. In the layout overlay options specify WALLDISPLAY=none (or off?). The wall is normally drawn around each overlay layout space and covers the x/y/x2/y2 axes.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Thank you so much.
That solves the problem!
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
For other people to learn, could you share your SAS data set named "gap1_map"?
Thanks,
Ethan
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
What about a small sample of masked data set?
Learn how use the CAT functions in SAS to join values from multiple variables into a single value.
Find more tutorials on the SAS Users YouTube channel.
SAS Training: Just a Click Away
Ready to level-up your skills? Choose your own adventure.