- Home
- /
- Programming
- /
- Graphics
- /
- Re: PROC CHART TRANSPARENT BAR by SUBGROUP
- RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Hello. I have a question. I want to create a bar graph by subgroup. This below the code is working well, but I want to change the transparency of all AMR bars but not the color (AIR should be no transparent). Can anyone help with it? Thank you so much.
**************
SAS code;
GOPTIONS RESET=ALL DEVICE=PNG TARGET=PNG HSIZE=8in VSIZE=7in FTITLE='Albany AMT/bold'
FTEXT='Albany AMT/bold' HTITLE=1.75 HTEXT=1.5;
data mileage;
input age & $15. sex $ amount sam & $15.;
call execute(catt('pattern',_N_,'color='('red','violet','BIGB','grey','STGB'),';');
datalines;
All sites M 500 , n=2812(100%)
All sites F 400 , n=1044(100%)
Female breast M 50 , n=554(20%)
Female breast F 5 , n=89(9%)
Male prostate M 110.0 , n=332(12%)
Male prostate F 10 , n=74(7%)
Lung & bronchus M 40 , n=242(9%)
Lung & bronchus F 35 , n=186(18%)
Colon & rectum M 45 , n=231(8%)
Colon & rectum F 70 , n=102(10%)
;
run;
data convert;
set mileage;
if sex='M' then amount=-amount;
run;
proc format;
picture posval low-high='000,009';
run;
data anlabels(drop=age sex );
length text color $ 50;
retain function 'label' when 'a' xsys ysys '2' hsys '3' size 2.5;
set convert;
midpoint=age; subgroup=sex;
text=strip(""||strip(left(put(amount,posval.)))||left(sam));
/*if age='All sites' then color='red';*/
/*else if age='Female breast' then color='violet';*/
/*else if age='Male prostate' then color='BIGB';*/
/*else if age='Lung & bronchus' then color='grey';*/
/*else color='STGB'; */
/* */
/* POSITION changes to place label outside of bar if bar is too small. */
if amount ge -115 and sex='M' then position='<';
else if sex='M' then position='>';
/* POSITION changes to place label outside of bar if bar is too small. */
if amount le 115 and sex='F' then position='>';
else if sex='F' then position='<';
output;
run;
/* Add a title to the graph */
title1 '';
axis1 label=(justify=left '') style=0 value=(f="Albany AMT/bold" h=1.5);
axis2 label=none value=( h=10pt) minor=none major=none
width=3 order=(-440 to 160 by 40) value=(f="Albany AMT/bold" h=1);
pattern1 v=solid color=red;
pattern2 v=solid color=STGB;
pattern3 v=solid color=violet;
pattern4 v=solid color=grey;
pattern5 v=solid color=BIGB;
/*pattern1 color = white;*/
/*pattern2 color = white;*/
proc gchart data=convert;
format amount posval.;
note font="Albany AMT/bold" move=(22pct,65pct) h=12pt 'AIR'
move=(80pct,65pct) 'AMR';
hbar age / sumvar=amount discrete nostat subgroup=sex SPACE =1 WIDTH =2
maxis=axis1 raxis=axis2 nolegend annotate=anlabels patternid=subgroup
patternid=midpoint ;
run;
quit;
Accepted Solutions
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
In that case, your best option is to create a variable with values that represent the unique combinations of age and sex (you can use 1, 2, 3, etc.), and use that as the GROUP variable. Then, use the attrmap technique that @Reeza showed, specifying the correct color and transparency for each combination value. Be sure to specify the NOAUTOLEGEND on the proc, as the legend will be pretty useless in this case.
Hope this helps!
Dan
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Sgplot is fine, too, How would you create? Thank you.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Your code errors out/doesn't work for me to create the sample data and/or the pattern statements.
71 data mileage; 72 input age & $15. sex $ amount sam & $15.; 73 74 datalines; NOTE: Invalid data for amount in line 76 5-9. RULE: ----+----1----+----2----+----3----+----4----+----5----+----6----+----7----+----8----+----9----+----0 76 All sites F 400 , n=1044(100%) age=All sites M 500 sex=All amount=. sam=F 400 , n=1044( _ERROR_=1 _N_=1 NOTE: Invalid data for amount in line 78 8-13. 78 Female breast F 5 , n=89(9%) age=Female breast M sex=Female amount=. sam=F 5 , n=89(9%) _ERROR_=1 _N_=2 NOTE: Invalid data for amount in line 80 6-13. 80 Male prostate F 10 , n=74(7%) age=Male prostate M sex=Male amount=. sam=F 10 , n=74(7%) _ERROR_=1 _N_=3 NOTE: Invalid data for amount in line 82 6-6. 82 Lung & bronchus F 35 , n=186(18%) age=Lung & bronchus sex=Lung amount=. sam=bronchus F 35 , _ERROR_=1 _N_=4 NOTE: Invalid data for amount in line 84 7-7. 84 Colon & rectum F 70 , n=102(10%) age=Colon & rectum sex=Colon amount=. sam=rectum F 70 , n _ERROR_=1 _N_=5 NOTE: SAS went to a new line when INPUT statement reached past the end of a line. NOTE: The data set WORK.MILEAGE has 5 observations and 4 variables.
You can use discrete attribute maps to specify the patterns, colours and shadings for specif groups. Here's an example from the documentation that controls the colours per groups.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Sorry this should work:
GOPTIONS RESET=ALL DEVICE=PNG TARGET=PNG HSIZE=8in VSIZE=7in FTITLE='Albany AMT/bold'
FTEXT='Albany AMT/bold' HTITLE=1.75 HTEXT=1.5;
data mileage;
input age & $15. sex $ amount sam & $15.;
datalines;
All sites M 500 , n=2812(100%)
All sites F 400 , n=1044(100%)
Female breast M 50 , n=554(20%)
Female breast F 5 , n=89(9%)
Male prostate M 110.0 , n=332(12%)
Male prostate F 10 , n=74(7%)
Lung & bronchus M 40 , n=242(9%)
Lung & bronchus F 35 , n=186(18%)
Colon & rectum M 45 , n=231(8%)
Colon & rectum F 70 , n=102(10%)
;
run;
data convert;
set mileage;
if sex='M' then amount=-amount;
run;
proc format;
picture posval low-high='000,009';
run;
data anlabels(drop=age sex );
length text color $ 50;
retain function 'label' when 'a' xsys ysys '2' hsys '3' size 2.5;
set convert;
midpoint=age; subgroup=sex;
text=strip(""||strip(left(put(amount,posval.)))||left(sam));
/*if age='All sites' then color='red';*/
/*else if age='Female breast' then color='violet';*/
/*else if age='Male prostate' then color='BIGB';*/
/*else if age='Lung & bronchus' then color='grey';*/
/*else color='STGB'; */
/* */
/* POSITION changes to place label outside of bar if bar is too small. */
if amount ge -115 and sex='M' then position='<';
else if sex='M' then position='>';
/* POSITION changes to place label outside of bar if bar is too small. */
if amount le 115 and sex='F' then position='>';
else if sex='F' then position='<';
output;
run;
/* Add a title to the graph */
title1 '';
axis1 label=(justify=left '') style=0 value=(f="Albany AMT/bold" h=1.5);
axis2 label=none value=( h=10pt) minor=none major=none
width=3 order=(-440 to 160 by 40) value=(f="Albany AMT/bold" h=1);
pattern1 v=solid color=red;
pattern2 v=solid color=STGB;
pattern3 v=solid color=violet;
pattern4 v=solid color=grey;
pattern5 v=solid color=BIGB;
/*pattern1 color = white;*/
/*pattern2 color = white;*/
proc gchart data=convert;
format amount posval.;
note font="Albany AMT/bold" move=(22pct,65pct) h=12pt 'AIR'
move=(80pct,65pct) 'AMR';
hbar age / sumvar=amount discrete nostat subgroup=sex SPACE =1 WIDTH =2
maxis=axis1 raxis=axis2 nolegend annotate=anlabels patternid=subgroup
patternid=midpoint ;
run;
quit;
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
**Please try this;
GOPTIONS RESET=ALL DEVICE=PNG TARGET=PNG HSIZE=8in VSIZE=7in FTITLE='Albany AMT/bold'
FTEXT='Albany AMT/bold' HTITLE=1.75 HTEXT=1.5;
data bars;
input cancer & $15. type $ rate sample & $15.;
datalines;
All sites I 423.8 , n=2812(100%)
All sites M 155.8 , n=1044(100%)
Female breast I 57.5 , n=554(20%)
Female breast M 23.2 , n=89(9%)
Male prostate I 110.0 , n=332(12%)
Male prostate M 29.2 , n=74(7%)
Lung & bronchus I 37.7 , n=242(9%)
Lung & bronchus M 27.7 , n=186(18%)
Colon & rectum I 35.6 , n=231(8%)
Colon & rectum M 15.8 , n=102(10%)
;
run;
data convert;
set bars;
if type='I' then rate=-rate;
run;
proc format;
picture posval low-high='000,009';
run;
data anlabels(drop=age sex );
length text color $ 50;
retain function 'label' when 'a' xsys ysys '2' hsys '3' size 2;
set convert;
midpoint=cancer; subgroup=type;
text=(" "||strip(left(put(rate,posval.)))||left(sample));
/*if age='All sites' then color='red';*/
/*else if age='Female breast' then color='violet';*/
/*else if age='Male prostate' then color='BIGB';*/
/*else if age='Lung & bronchus' then color='grey';*/
/*else color='STGB'; */
/* */
/* POSITION changes to place label outside of bar if bar is too small. */
if rate ge -115 and type='I' then position='<';
else if type='I' then position='>';
/* POSITION changes to place label outside of bar if bar is too small. */
if rate le 115 and type='M' then position='>';
else if type='M' then position='<';
output;
run;
/* Add a title to the graph */
title1 '';
axis1 label=(justify=left '') style=0 value=(f="Albany AMT/bold" h=1);
axis2 label=none value=( h=10pt) minor=none major=none
width=3 order=(-440 to 160 by 40) value=(f="Albany AMT/bold" h=1);
pattern1 v=solid color=red;
pattern2 v=solid color=STGB;
pattern3 v=solid color=violet;
pattern4 v=solid color=grey;
pattern5 v=solid color=BIGB;
/*pattern1 color = white;*/
/*pattern2 color = white;*/
proc gchart data=convert;
format rate posval.;
note font="Albany AMT/bold" move=(22pct,65pct) h=12pt 'AIR'
move=(80pct,65pct) 'AMR';
hbar cancer / sumvar=rate discrete nostat subgroup=type SPACE =1 WIDTH =2
maxis=axis1 raxis=axis2 nolegend annotate=anlabels patternid=subgroup
patternid=midpoint ;
run;
quit;
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
data work.mileage; input age & $15. sex $ amount sam & $15.; datalines; All sites M 500 n=2812(100%) All sites F 400 n=1044(100%) Female breast M 50 n=554(20%) Female breast F 5 n=89(9%) Male prostate M 110.0 n=332(12%) Male prostate F 10 n=74(7%) Lung & bronchus M 40 n=242(9%) Lung & bronchus F 35 n=186(18%) Colon & rectum M 45 n=231(8%) Colon & rectum F 70 n=102(10%) ;
The forum message windows will reformat text, typically removing white space such as reducing two spaces to one. So code pasted into the main message windows often does not run quite right when reading some forms of data. Paste code into a code box opened with the {I} or "running man" icon to preserve formatting.
It really does not help to change the names of data sets, variables or values from example to example.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Sorry, attached the excel data. Does it work? Thank you.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Paste the text into the code is much easier to work with, see BallardW example or this one.
It's the 6/7th icon in the editor window and it will maintain formatting.
This should get you started. You'll have to still figure out how to change the bar colours, which can also be done via styleattrs statement or directly in the plot. To get the labels in, instead of annotation data set you can use something very similar which is a TEXT statement in the graph. SGPLOT is much easier to customize IMO and you get better quality graphics.
But it's also a learning curve.
data attrmaptrans;
retain linecolor "black";
input id $ value $ fillcolor $ filltransparency;
datalines;
sex F red 0
sex M green 0.7
;
run;
title 'New Graph';
proc sgplot data=convert dattrmap = attrmaptrans;
hbarbasic age / stat=sum
response=amount
group=sex
attrid=sex
dataskin=gloss;
format amount posval.;
text text=sam x= amount y= age;
label sex = "Sex";
run;- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Sorry, that is not what I wanted. I want each age group colored differently and by gender transparent for males and not transparent for female.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
In that case, your best option is to create a variable with values that represent the unique combinations of age and sex (you can use 1, 2, 3, etc.), and use that as the GROUP variable. Then, use the attrmap technique that @Reeza showed, specifying the correct color and transparency for each combination value. Be sure to specify the NOAUTOLEGEND on the proc, as the legend will be pretty useless in this case.
Hope this helps!
Dan
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
@Zula wrote:
Sorry, that is not what I wanted. I want each age group colored differently and by gender transparent for males and not transparent for female.
You're correct, I did not take the time to replicate your entire graph. You can extend it using the method Rick mentioned and would still need to customize your fonts, spacing and sizes to what you need.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
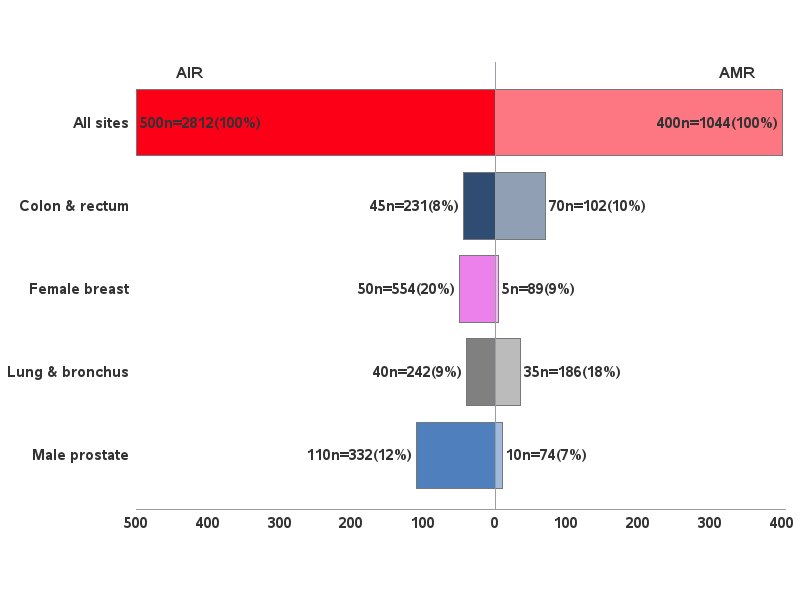
Here's a way to do it with Gchart ...
data my_data;
length category $20 subgroup $10 sam $20 colorvar $20;
infile datalines dlm=',';
input category subgroup amount sam;
colorvar=trim(left(subgroup))||'_'||trim(left(category));
datalines;
All sites,AIR,500,n=2812(100%)
All sites,AMR,400,n=1044(100%)
Female breast,AIR,50,n=554(20%)
Female breast,AMR,5,n=89(9%)
Male prostate,AIR,110.0,n=332(12%)
Male prostate,AMR,10,n=74(7%)
Lung & bronchus,AIR,40,n=242(9%)
Lung & bronchus,AMR,35,n=186(18%)
Colon & rectum,AIR,45,n=231(8%)
Colon & rectum,AMR,70,n=102(10%)
;
run;
proc format;
picture posval low-high='000,009';
run;
data my_data; set my_data;
if subgroup='AIR' then amount=-amount;
run;
data anlabels; set my_data;
length text $100;
xsys='2'; ysys='2'; hsys='3'; when='a';
yc=category;
x=amount;
function='label'; size=2.5;
text=strip(""||strip(left(put(amount,posval.)))||left(sam));
if subgroup='AMR' then do;
text='a0'x||trim(left(text));
position='>';
end;
if subgroup='AIR' then do;
text=trim(left(text))||'a0'x;
position='<';
end;
if category='All sites' and subgroup='AMR' then do;
text=trim(left(text))||'a0'x;
position='<';
end;
if category='All sites' and subgroup='AIR' then do;
text='a0'x||trim(left(text));
position='>';
end;
run;
goptions device=png;
goptions gunit=pct htitle=18pt ftitle="albany amt/bold" htext=12pt ftext="albany amt/bold";
goptions ctext=gray33;
axis1 label=none value=(height=11pt font="Albany AMT/bold" justify=right)
style=0 offset=(9,10);
axis2 label=none value=(height=11pt font="Albany AMT/bold")
minor=none major=none /*width=3*/;
legend1 label=none shape=bar(.15in,.15in) across=2 colmajor;
/* solid colors for the AIR bars */
pattern1 v=solid color=cxfc0018;
pattern2 v=solid color=cx2f4c72;
pattern3 v=solid color=cxec81ec;
pattern4 v=solid color=cx808080;
pattern5 v=solid color=cx4f7fbd;
/* transparent colors for the AMR bars */
pattern6 v=solid color=Afc001888;
pattern7 v=solid color=A2f4c7288;
pattern8 v=solid color=Aec81ec88;
pattern9 v=solid color=A80808088;
pattern10 v=solid color=A4f7fbd88;
title1;
footnote;
proc gchart data=my_data;
format amount posval.;
note font="Albany AMT/bold" h=12pt
move=(22pct,87pct) 'AIR'
move=(90pct,87pct) 'AMR';
hbar category / sumvar=amount nostat
maxis=axis1 raxis=axis2
width=6 space=1.5 coutline=gray77
annotate=anlabels
subgroup=colorvar
nolegend /* legend=legend1 */;
run;
Learn how use the CAT functions in SAS to join values from multiple variables into a single value.
Find more tutorials on the SAS Users YouTube channel.
SAS Training: Just a Click Away
Ready to level-up your skills? Choose your own adventure.