- Home
- /
- Programming
- /
- Graphics
- /
- How to avoid the datalabels overlapping on Error bars in Forest Plot
- RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
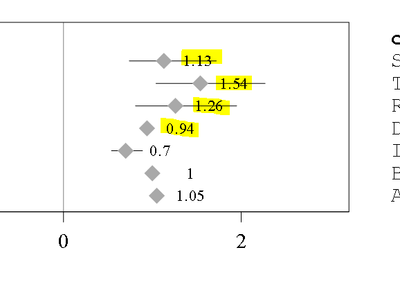
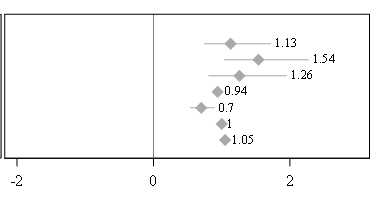
I am creating the data labels on the forest plot. However My data labels overlapping with the Error bars. How I can Avoid this. I tried using DATALABELPOSITION But no luck. I thought of Creating Blank space before dataset ( data point values). However it won't change it dynamically , if any values changes in future. Can you please suggest the Options.
Example: How the labels should look!
How My labels are displaying
layout overlay /xaxisopts=(label=' ' linearopts =(tickvaluelist=(-2 0 2) viewmin=-2 viewmax=2))
/*type=discrete discreteopts =( tickvaluelist=( '-0.5' '0' '0.5' '1' '2')))*/
y2axisopts = (display=none);
scatterplot x=OR y=ORCI /
markerattrs = (symbol=diamondfilled size=0 color=white) yaxis=y2;
scatterplot x=OR y=ORCI / datalabel = OR datalabelattrs = (size=7) datalabelposition=right
markerattrs = (symbol=diamondfilled size=10 color=darkgrey)
yaxis=y2
xerrorlower = lcl xerrorupper = ucl errorbarcapshape = none;
referenceline x=0;
annotate;
endlayout;
Accepted Solutions
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Instead using the error bar on the SCATTERPLOT statement, use a HIGHLOWPLOT plot. Set the LOWCAP=NONE and HIGHCAP=none, and set the HIGHLABEL to be the same variable as the DATALABEL. The change should look something like the following:
layout overlay /xaxisopts=(label=' ' linearopts =(tickvaluelist=(-2 0 2) viewmin=-2 viewmax=2))
/*type=discrete discreteopts =( tickvaluelist=( '-0.5' '0' '0.5' '1' '2')))*/
y2axisopts = (display=none);
scatterplot x=OR y=ORCI / markerattrs = (symbol=diamondfilled size=0 color=white) yaxis=y2;
highlowplot y=ORCI low=lcl high=ucl / lowcap=none highcap=none highlabel=OR labelattrs=(size=7)
lineattrs=(color=darkgrey) yaxis=y2;
scatterplot x=OR y=ORCI /
markerattrs = (symbol=diamondfilled size=10 color=darkgrey)
yaxis=y2;
referenceline x=0;
annotate;
endlayout;Hope this helps!
Dan
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Instead using the error bar on the SCATTERPLOT statement, use a HIGHLOWPLOT plot. Set the LOWCAP=NONE and HIGHCAP=none, and set the HIGHLABEL to be the same variable as the DATALABEL. The change should look something like the following:
layout overlay /xaxisopts=(label=' ' linearopts =(tickvaluelist=(-2 0 2) viewmin=-2 viewmax=2))
/*type=discrete discreteopts =( tickvaluelist=( '-0.5' '0' '0.5' '1' '2')))*/
y2axisopts = (display=none);
scatterplot x=OR y=ORCI / markerattrs = (symbol=diamondfilled size=0 color=white) yaxis=y2;
highlowplot y=ORCI low=lcl high=ucl / lowcap=none highcap=none highlabel=OR labelattrs=(size=7)
lineattrs=(color=darkgrey) yaxis=y2;
scatterplot x=OR y=ORCI /
markerattrs = (symbol=diamondfilled size=10 color=darkgrey)
yaxis=y2;
referenceline x=0;
annotate;
endlayout;Hope this helps!
Dan
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Perfect. It worked. Thanks Dan.😍
Learn how use the CAT functions in SAS to join values from multiple variables into a single value.
Find more tutorials on the SAS Users YouTube channel.
SAS Training: Just a Click Away
Ready to level-up your skills? Choose your own adventure.