- Home
- /
- Programming
- /
- Graphics
- /
- Re: GTL Layout Lattice and Forest Plot - remove gridlines and remove f...
- RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
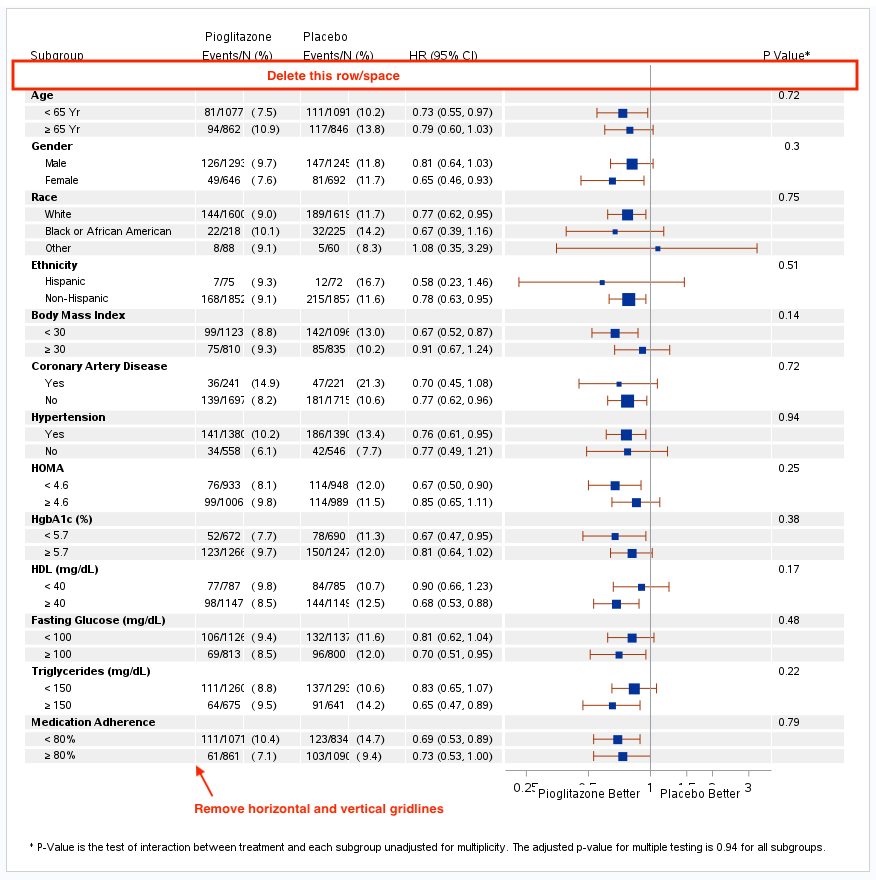
Using this forest plot example from https://www.pharmasug.org/proceedings/2017/QT/PharmaSUG-2017-QT15.pdf, how do I remove the white horizontal and vertical gridlines and remove the first row/blank space in the table?
I attempted to remove the gridlines by inserting the following snippet of code after the first layout lattice statement, but it did not work:
rowaxes;
rowaxis /griddisplay=off;
endrowaxes;
Here is the code used to generate the forest plot:
%let dpi=300;
%let w=9in;
%let h=6.5in;
data SubgroupsData;
infile datalines delimiter=',';
input Indent Subgroup : $3-27 CountA : best12. EventA : best12. CountB : best12. EventB : best12. HR CIL CIU PValue : 7.2;
format EventDisplayA $4. CountDisplayA $4. EventDisplayB $4. CountDisplayB $4. ;
if CountA ne . then do;
PctA = (EventA/CountA)*100;
PctB = (EventB/CountB)*100;
EventDisplayA = right(put(EventA, 4.0));
CountDisplayA = left(put(CountA, 4.0));
EventCountA = right(EventDisplayA || "/" || CountDisplayA);
PctDisplayA = "(" || put(PctA, 4.1) || ")";
EventDisplayB = right(put(EventB, 4.0));
CountDisplayB = left(put(CountB, 4.0));
EventCountB = right(EventDisplayB || "/" || CountDisplayB);
PctDisplayB = "(" || put(PctB, 4.1) || ")";
HRCI = put(HR, 4.2) || " (" || put(CIL, 4.2) || ", "
|| put(CIU, 4.2) || ")";
/* Determine the marker size based on population size */
SquareSize = ((CountA + CountB)/3876) * 12 ;
end;
row = _n_;
datalines;
0,Age,.,.,.,.,.,.,.,0.72
1,< 65 Yr,1077,81,1091,111,0.73,0.55,0.97,.
1,>= 65 Yr,862,94,846,117,0.79,0.60,1.03,.
0,Gender,.,.,.,.,.,.,.,0.30
1,Male,1293,126,1245,147,0.81,0.64,1.03,.
1,Female,646,49,692,81,0.65,0.46,0.93,.
0,Race,.,.,.,.,.,.,.,0.75
1,White,1600,144,1619,189,0.77,0.62,0.95,.
1,Black or African American,218,22,225,32,0.67,0.39,1.16,.
1,Other,88,8,60,5,1.08,0.35,3.29,.
0,Ethnicity,.,.,.,.,.,.,.,0.51
1,Hispanic,75,7,72,12,0.58,0.23,1.46,.
1,Non-Hispanic,1852,168,1857,215,0.78,0.63,0.95,.
0,Body Mass Index,.,.,.,.,.,.,.,0.14
1,< 30,1123,99,1096,142,0.67,0.52,0.87,.
1,>= 30,810,75,835,85,0.91,0.67,1.24,.
0,Coronary Artery Disease,.,.,.,.,.,.,.,0.72
1,Yes,241,36,221,47,0.70,0.45,1.08,.
1,No,1697,139,1715,181,0.77,0.62,0.96,.
0,Hypertension,.,.,.,.,.,.,.,0.94
1,Yes,1380,141,1390,186,0.76,0.61,0.95,.
1,No,558,34,546,42,0.77,0.49,1.21,.
0,HOMA,.,.,.,.,.,.,.,0.25
1,< 4.6,933,76,948,114,0.67,0.50,0.90,.
1,>= 4.6,1006,99,989,114,0.85,0.65,1.11,.
0,HgbA1c (%),.,.,.,.,.,.,.,0.38
1,< 5.7,672,52,690,78,0.67,0.47,0.95,.
1,>= 5.7,1266,123,1247,150,0.81,0.64,1.02,.
0,HDL (mg/dL),.,.,.,.,.,.,.,0.17
1,< 40,787,77,785,84,0.90,0.66,1.23,.
1,>= 40,1147,98,1149,144,0.68,0.53,0.88,.
0,Fasting Glucose (mg/dL),.,.,.,.,.,.,.,0.48
1,< 100,1126,106,1137,132,0.81,0.62,1.04,.
1,>= 100,813,69,800,96,0.70,0.51,0.95,.
0,Triglycerides (mg/dL),.,.,.,.,.,.,.,0.22
1,< 150,1260,111,1293,137,0.83,0.65,1.07,.
1,>= 150,675,64,641,91,0.65,0.47,0.89,.
0,Medication Adherence,.,.,.,.,.,.,.,0.79
1,< 80%,1071,111,834,123,0.69,0.53,0.89,.
1,>= 80%,861,61,1090,103,0.73,0.53,1.00,.
;
/*--Used for Subgroup labels in column 1--*/
data anno3(drop=indent);
set SubgroupsData(keep=row subgroup indent rename=(row=y1));
retain Function 'Text ' ID 'id1' X1Space 'DataPercent'
Y1Space 'DataValue ' x1 x2 2 TextSize 7 Width 100 Anchor 'Left ';
if indent;
label = tranwrd(subgroup, '>=', '(*ESC*){Unicode ''2265''x}');
run;
/*--Used for text under x axis of HR scatter plot in column 7--*/
data anno4;
retain Function 'Arrow' ID 'id2' X1Space X2Space 'DataValue'
FIllTransparency 0 Y1Space Y2Space 'GraphPercent'
Scale 1e-40 LineThickness 1 y1 4.0 y2 4.0 Width 100
FillStyleElement 'GraphWalls' LineColor 'Black';
y1=0;y2=100;x2=x1; function='Arrow'; output;
function = 'Text';
y1 = 4.0;
y2 = 4.0;
x1 = 0.92; anchor = 'Right'; label = 'Pioglitazone Better'; Textsize=8; output;
x1 = 1.07; anchor = 'Left '; label = 'Placebo Better'; Textsize=8; output;
run;
data anno5; set anno3 anno4; run;
data forest3(drop=flag);
set SubgroupsData nobs=nobs;
Head = not indent;
retain flag 0;
if head then flag = mod(flag + 1, 2);
if flag then ref=row;
if indent then subgroup = ' ';
run;
/*--Define template for Forest Plot--*/
/*--Template uses a Layout Lattice of 8 columns--*/
proc template;
define statgraph Forest3;
dynamic /*_show_bands*/ _color _thk;
begingraph;
discreteattrmap name='text';
value '1' / textattrs=(weight=bold); value other;
enddiscreteattrmap;
discreteattrvar attrvar=type var=head attrmap='text';
layout lattice /
columns=8 columnweights=(0.21 0.06 .07 0.06 .07 0.12 0.33 0.09);
/*--Column headers--*/
sidebar / align=top;
layout lattice /
rows=2 columns=5 columnweights=(0.21 0.12 0.13 0.40 0.14);
entry " ";
entry textattrs=(size=8) halign=left " Pioglitazone";
entry textattrs=(size=8) halign=left " Placebo";
entry " ";
entry " ";
entry textattrs=(size=8) halign=left "Subgroup";
entry textattrs=(size=8) halign=left "Events/N (%)";
entry textattrs=(size=8) halign=left " Events/N (%)";
entry textattrs=(size=8) halign=left " HR (95% CI)";
entry halign=center textattrs=(size=8) "P Value*" ;
endlayout;
endsidebar;
/*--First Subgroup column, shows only the Y2 axis--*/
layout overlay / walldisplay=none xaxisopts=(display=none)
yaxisopts=(reverse=true display=none
tickvalueattrs=(weight=bold));
annotate / id='id1';
referenceline y=ref / lineattrs=(thickness=_thk color=_color);
axistable y=row value=subgroup /
display=(values) textgroup=type;
endlayout;
/*--Second column showing Pioglitizone Events/Count --*/
layout overlay / xaxisopts=(display=none)
yaxisopts=(reverse=true display=none) walldisplay=none;
referenceline y=ref / lineattrs=(thickness=_thk color=_color);
axistable y=row value=EventCountA /display=(values)
valuejustify = center;
endlayout;
/*--Third column showing Pioglitizone Percent--*/
layout overlay / xaxisopts=(display=none)
yaxisopts=(reverse=true display=none) walldisplay=none;
referenceline y=ref / lineattrs=(thickness=_thk color=_color);
axistable y=row value=PctDisplayA /display=(values)
valuejustify = left;
endlayout;
/*--Fourth column showing Placebo Events/Count --*/
layout overlay / xaxisopts=(display=none)
yaxisopts=(reverse=true display=none) walldisplay=none;
referenceline y=ref / lineattrs=(thickness=_thk color=_color);
axistable y=row value=EventCountB /display=(values)
valuejustify = center;
endlayout;
/*--Fifth column showing Placebo Percent--*/
layout overlay / xaxisopts=(display=none)
yaxisopts=(reverse=true display=none) walldisplay=none;
referenceline y=ref / lineattrs=(thickness=_thk color=_color);
axistable y=row value=PctDisplayB /display=(values)
valuejustify = left;
endlayout;
/*--Sixth column showing PCIGroup--*/
layout overlay / x2axisopts=(display=none)
yaxisopts=(reverse=true display=none) walldisplay=none;
referenceline y=ref / lineattrs=(thickness=_thk color=_color);
axistable y=row value=HRCI / display=(values);
endlayout;
/*--Seventh column showing Hazard ratio graph with 95% error bars--*/
layout overlay / xaxisopts=(type=log
label=' '
labelattrs=(size=10)
logopts=(tickvaluepriority=true
tickvaluelist=(0.25 0.5 1.0 1.5 2.0 3.0)))
yaxisopts=(reverse=true display=none) walldisplay=none;
annotate / id='id2';
referenceline y=ref / lineattrs=(thickness=_thk color=_color);
scatterplot y=row x=HR / xerrorlower=CIL xerrorupper=CIU
sizeresponse=SquareSize sizemin=4 sizemax=12
markerattrs=(symbol=squarefilled);
referenceline x=1;
endlayout;
/*--Eigth column showing P-Values--*/
layout overlay / x2axisopts=(display=none)
yaxisopts=(reverse=true display=none) walldisplay=none;
referenceline y=ref / lineattrs=(thickness=_thk color=_color);
axistable y=row value=pvalue / display=(values)
valuejustify = right
showmissing=false; /*false removes . for missing pvalues*/
endlayout;
endlayout;
entryfootnote halign=left textattrs=(size=7)
'* P-Value is the test of interaction between treatment and each subgroup unadjusted for multiplicity. The adjusted '
'p-value for multiple testing is 0.94 for all subgroups.';
endgraph;
end;
run;
proc sgrender data=Forest3 template=Forest3 sganno=anno5;
dynamic _color='greyef' _thk=12 ;
run;
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
CTSPedia Clinical Graphs - Subgrouped Forest Plot
By Sanjay Matange on Graphically Speaking May 30, 2016
https://blogs.sas.com/content/graphicallyspeaking/2016/05/30/ctspedia-clinical-graphs-subgrouped-for...
Tips and tricks: Segmented discrete axis
By Sanjay Matange on Graphically Speaking October 22, 2017
https://blogs.sas.com/content/graphicallyspeaking/2017/10/22/tips-tricks-segmented-discrete-axis/
Koen
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
The apparent white horizontal gaps are due to the reference line of thickness=_thk(12) does not fill the space. If you have 9.4M5, you can use the
USEDISCRETETHICKNESS=TRUE and DISCRETELINETHICKNESS=1 to fill the space fully.
See: https://documentation.sas.com/doc/en/pgmsascdc/9.4_3.5/grstatgraph/p03jtr6n56wu4ln1k77szqw1fjkc.htm
The vertical lines white lines are the gaps between the columns that show because the grey color reflines do not draw across the column borders.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Hello @PharmlyDoc ,
To find out if you have Maintenance Level 5 or later, submit:
%put &=sysvlong4;For example, I have Maintenance Level 7:
1 %put &=sysvlong4; SYSVLONG4=9.04.01M7P08052020
Koen
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
1 %put &=sysvlong4;
SYSVLONG4=9.04.01M5P09132017
I forgot to add the height and width options that I used:
ods graphics / imagemap width=9in height=9in) ;
proc sgrender data=Forest3 template=Forest3 sganno=anno5;
dynamic _color='greyef' _thk=15 ;
run;I increased the line thickness to 15 which fixed the row grids --> _thk = 15.
Is the dynamic statement and its options interfering with the ability to remove the column gridlines?
Why doesn't the following code work?
columnaxes;
columnaxis /griddisplay=off;
endcolumnaxes;
Where would I input USEDISCRETETHICKNESS=TRUE DISCRETELINETHICKNESS=1 ?
How do I remove the column gridlines or make the reflines draw across the column borders?
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
While setting _thk=15, may work for now, this may fail if the data changes. Better to use the USEDISCRETETHICKNESS=TRUE DISCRETELINETHICKNESS=1 in the REFERENCELINE statement. See previous link provided.
As I said before, the vertical white lines are NOT gridlines. This is the gap between the columns of your layout. The horizontal gray reference lines do not draw in the column gap. I don't think there is an easy way to fix this by any options.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Hi,my friends:
when i rerun your code,in the data anno3 and forest3 steps,add the code 'if row>37 then delete in both two data step' ,then the output forest plot appear abnormal ,two long blank will display in the bottow of the forest,wheather you and others had found the same ?
Learn how use the CAT functions in SAS to join values from multiple variables into a single value.
Find more tutorials on the SAS Users YouTube channel.
SAS Training: Just a Click Away
Ready to level-up your skills? Choose your own adventure.