- Home
- /
- Programming
- /
- Programming
- /
- Re: Interval estimation for binomial proportion: Wilson interval
- RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
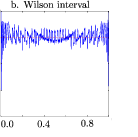
I need code for coverage probability (Wilson interval) to get following graph
thank you
P.S.
i found macro, but it didnt work for me.
Calculating the coverage probability for a given N and binomial p can be done in a single data step, summing the probability-weighted coverage indicators over the realized values of the random variate. Once this machinery is developed, we can call it repeatedly, using a macro, to find the results for different binomial p. We comment on the code internally.
%macro onep(n=,p=,alpha=.05);
data onep;
n = &n;
p = &p;
alpha = α
/* set up collectors of the weighted coverage indicators */
expcontrib_t = 0;
expcontrib_p4 = 0;
expcontrib_s = 0;
expcontrib_cp = 0;
/* loop through the possible observed successes x*/
do x = 0 to n;
probobs = pdf('BINOM',x,p,n); /* probability X=x */
phat = x/n;
zquant = quantile('NORMAl', 1 - alpha/2, 0, 1);
p4 = (x+2)/(n + 4);
/* calculate the half-width of the t and plus4 intervals */
thalf = quantile('T', 1 - alpha/2,(n-1)) * sqrt(phat*(1-phat)/n);
p4half = zquant * sqrt(p4*(1-p4)/(n+4));
/* the score CI in R uses a Yates correction by default, and is
reproduced here */
yates = min(0.5, abs(x - (n*p)));
z22n = (zquant**2)/(2*n);
yl = phat-yates/n;
yu = phat+yates/n;
slower = (yl + z22n - zquant * sqrt( (yl*(1-yl)/n) + z22n / (2*n) )) /
(1 + 2 * z22n);
supper = (yu + z22n + zquant * sqrt( (yu*(1-yu)/n) + z22n / (2*n) )) /
(1 + 2 * z22n);
/* cover = 1 if in the CI, 0 else */
cover_t = ((phat - thalf) < p) and ((phat + thalf) > p);
cover_p4 = ((p4 - p4half) < p) and ((p4 + p4half) > p);
cover_s = (slower < p) and (supper > p);
/* the Clopper-Pearson interval can be easily calculated on the fly */
cover_cp = (quantile('BETA', alpha/2 ,x,n-x+1) < p) and
(quantile('BETA', 1 - alpha/2 ,x+1,n-x) > p);
/* cumulate the weighted probabilities */
expcontrib_t = expcontrib_t + probobs * cover_t;
expcontrib_p4 = expcontrib_p4 + probobs * cover_p4;
expcontrib_s = expcontrib_s + probobs * cover_s;
expcontrib_cp = expcontrib_cp + probobs * cover_cp;
/* only save the last interation */
if x = N then output;
end;
run;
%mend onep;
The following macro calls the first one for a series of binomial p for a fixed N. Since the macro %do loop can only iterate through integers, we have to do a little division; the %sysevelf function will do this within the macro.
%macro repcicov(n=, lop=, hip=, byp=, alpha= .05);
/* need an empty data set to store the results */
data summ; set _null_; run;
%do stepp = %sysevalf(&lop / &byp, floor) %to %sysevalf(&hip / &byp,floor);
/* note that the p sent to the %onep macro is a
text string like "49 * .001" */
%onep(n = &n, p = &stepp * &byp, alpha = &alpha);
/* tack on the current results to the ones finished so far */
/* this is a simple but inefficient way to add each binomial p into
the output data set */
data summ; set summ onep; run;
%end;
%mend repcicov;
/* same parameters as in R */
%repcicov(n=50, lop = .01, hip = .3, byp = .001);
Finally, we can plot the results. One option shown here and not mentioned in the book are the mode=include option to the symbol statement, which allows the two distinct pieces of the T coverage to display correctly.
goptions reset=all;
legend1 label=none position=(bottom right inside)
mode=share across=1 frame value = (h=2);
axis1 order = (0.85 to 1 by 0.05) minor=none
label = (a=90 h=2 "Coverage probability") value=(h=2);
axis2 order = (0 to 0.3 by 0.05) minor=none
label = (h=2 "True binomial p") value=(h=2);
symbol1 i = j v = none l =1 w=3 c=blue mode=include;
symbol2 i = j v = none l =1 w=3 c=red;
symbol3 i = j v = none l =1 w=3 c=lightgreen;
symbol4 i = j v = none l =1 w=3 c=black;
proc gplot data = summ;
plot (expcontrib_t expcontrib_p4 expcontrib_s expcontrib_cp) * p
/ overlay legend vaxis = axis1 haxis = axis2 vref = 0.95 legend = legend1;
label expcontrib_t = "T approximation" expcontrib_p4 = "P4 method"
expcontrib_s = "Score method" expcontrib_cp = "Exact (CP)";
run; quit;
Accepted Solutions
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Hello @bigban777,
I haven't checked all the details of the code you posted, but after the following corrections/modifications, the graphs are produced and look plausible:
- Line 5 must read: alpha = α
- The calculation of cover_cp fails in the degenerate cases x=0 and x=n, so these cases need to be handled separately:
if 0<x<n then cover_cp = (quantile('BETA', alpha/2 ,x,n-x+1) < p) and (quantile('BETA', 1 - alpha/2 ,x+1,n-x) > p); else if x=0 then cover_cp = (0 < p) and (quantile('BETA', 1 - alpha/2 ,x+1,n-x) > p); else if x=n then cover_cp = (quantile('BETA', alpha/2 ,x,n-x+1) < p) and (1 > p); - Of course, the parameters in the call of macro REPCICOV have to be adjusted to the n and p range you want. The latter appears to be the interval [0, 1] in your graph. So, you could use
(Please choose the value of n yourself.)%repcicov(n=50, lop = 0, hip = 1, byp = .001); - The x axis range needs to be adapted accordingly:
axis2 order = (0 to 1 by 0.05) minor=none - If you want to see the graph for the Wilson interval alone, you can delete the other three plot requests or comment them out:
plot (/* expcontrib_t expcontrib_p4 */ expcontrib_s /* expcontrib_cp */) * p
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Hello @bigban777,
I haven't checked all the details of the code you posted, but after the following corrections/modifications, the graphs are produced and look plausible:
- Line 5 must read: alpha = α
- The calculation of cover_cp fails in the degenerate cases x=0 and x=n, so these cases need to be handled separately:
if 0<x<n then cover_cp = (quantile('BETA', alpha/2 ,x,n-x+1) < p) and (quantile('BETA', 1 - alpha/2 ,x+1,n-x) > p); else if x=0 then cover_cp = (0 < p) and (quantile('BETA', 1 - alpha/2 ,x+1,n-x) > p); else if x=n then cover_cp = (quantile('BETA', alpha/2 ,x,n-x+1) < p) and (1 > p); - Of course, the parameters in the call of macro REPCICOV have to be adjusted to the n and p range you want. The latter appears to be the interval [0, 1] in your graph. So, you could use
(Please choose the value of n yourself.)%repcicov(n=50, lop = 0, hip = 1, byp = .001); - The x axis range needs to be adapted accordingly:
axis2 order = (0 to 1 by 0.05) minor=none - If you want to see the graph for the Wilson interval alone, you can delete the other three plot requests or comment them out:
plot (/* expcontrib_t expcontrib_p4 */ expcontrib_s /* expcontrib_cp */) * p
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Hi,
i did change in my code as you said but it doesnt show graph. Here is my fixes code. Where is mistake?
%macro onep(n=,p=,alpha=.05);
data onep;
n = &n;
p = &p;
alpha = α
/* set up collectors of the weighted coverage indicators */
expcontrib_t = 0;
expcontrib_p4 = 0;
expcontrib_s = 0;
expcontrib_cp = 0;
/* loop through the possible observed successes x*/
do x = 0 to n;
probobs = pdf('BINOM',x,p,n); /* probability X=x */
phat = x/n;
zquant = quantile('NORMAl', 1 - alpha/2, 0, 1);
p4 = (x+2)/(n + 4);
/* calculate the half-width of the t and plus4 intervals */
thalf = quantile('T', 1 - alpha/2,(n-1)) * sqrt(phat*(1-phat)/n);
p4half = zquant * sqrt(p4*(1-p4)/(n+4));
/* the score CI in R uses a Yates correction by default, and is
reproduced here */
yates = min(0.5, abs(x - (n*p)));
z22n = (zquant**2)/(2*n);
yl = phat-yates/n;
yu = phat+yates/n;
slower = (yl + z22n - zquant * sqrt( (yl*(1-yl)/n) + z22n / (2*n) )) /
(1 + 2 * z22n);
supper = (yu + z22n + zquant * sqrt( (yu*(1-yu)/n) + z22n / (2*n) )) /
(1 + 2 * z22n);
/* cover = 1 if in the CI, 0 else */
cover_t = ((phat - thalf) < p) and ((phat + thalf) > p);
cover_p4 = ((p4 - p4half) < p) and ((p4 + p4half) > p);
cover_s = (slower < p) and (supper > p);
/* the Clopper-Pearson interval can be easily calculated on the fly */
cover_cp = (quantile('BETA', alpha/2 ,x,n-x+1) < p) and
(quantile('BETA', 1 - alpha/2 ,x+1,n-x) > p);
if 0<x<n then
cover_cp = (quantile('BETA', alpha/2 ,x,n-x+1) < p) and
(quantile('BETA', 1 - alpha/2 ,x+1,n-x) > p);
else if x=0 then
cover_cp = (0 < p) and (quantile('BETA', 1 - alpha/2 ,x+1,n-x) > p);
else if x=n then
cover_cp = (quantile('BETA', alpha/2 ,x,n-x+1) < p) and (1 > p);
/* cumulate the weighted probabilities */
expcontrib_t = expcontrib_t + probobs * cover_t;
expcontrib_p4 = expcontrib_p4 + probobs * cover_p4;
expcontrib_s = expcontrib_s + probobs * cover_s;
expcontrib_cp = expcontrib_cp + probobs * cover_cp;
/* only save the last interation */
if x = N then output;
end;
run;
%mend onep;
%macro repcicov(n=, lop=, hip=, byp=, alpha= .05);
/* need an empty data set to store the results */
data summ; set _null_; run;
%do stepp = %sysevalf(&lop / &byp, floor) %to %sysevalf(&hip / &byp,floor);
/* note that the p sent to the %onep macro is a
text string like "49 * .001" */
%onep(n = &n, p = &stepp * &byp, alpha = &alpha);
/* tack on the current results to the ones finished so far */
/* this is a simple but inefficient way to add each binomial p into
the output data set */
data summ; set summ onep; run;
%end;
%mend repcicov;
/* same parameters as in R */
%repcicov(n=50, lop = 0, hip = 1, byp = .001);
goptions reset=all;
legend1 label=none position=(bottom right inside)
mode=share across=1 frame value = (h=2);
axis1 order = (0.85 to 1 by 0.05) minor=none
label = (a=90 h=2 "Coverage probability") value=(h=2);
axis2 order = (0 to 0.3 by 0.05) minor=none
label = (h=2 "True binomial p") value=(h=2);
symbol1 i = j v = none l =1 w=3 c=blue mode=include;
symbol2 i = j v = none l =1 w=3 c=red;
symbol3 i = j v = none l =1 w=3 c=lightgreen;
symbol4 i = j v = none l =1 w=3 c=black;
proc gplot data = summ;
plot (/*expcontrib_t expcontrib_p4 */ expcontrib_s /*expcontrib_cp*/) * p
/ overlay legend vaxis = axis1 haxis = axis2 vref = 0.95 legend = legend1;
label expcontrib_t = "T approximation" expcontrib_p4 = "P4 method"
expcontrib_s = "Score method" expcontrib_cp = "Exact (CP)";
run; quit;Thank you
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
@bigban777 wrote:
Hi,
i did change in my code as you said
Well, you did, but neither correctly, nor completely.
- The new definition of cover_cp should have replaced the old one. So, just delete the two lines of code after the comment
"/* the Clopper-Pearson interval ...". - You forgot to implement item 4: axis2 order = (0 to 1 by 0.05) minor=none
Please note that this axis definition is to replace the old axis2 statement.
But even without these corrections your code produces a (partial) graph in my SAS session. Maybe you have to double-click the item representing the plot in the results window?
[Edit 2019-04-30: Reattached screenshot, which had been deleted by mistake.]
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
for some reason i couldnt get result. I use SAS on Demand.
Could you post graph what you got please.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
I am sorry, but I think this might violate the terms of my SAS license agreement.
I'm not familiar with SAS on Demand. Are you unable to create any graph with that software?
If so, I suggest you post this general problem in a new thread, e.g., in the "SAS/GRAPH and ODS Graphics" subforum.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Is this the same thing PROC FREQ can do?
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
@data_null__: Yes, I assume it's the same CI (but haven't checked). However, I guess it would not help much to replace the data step formula in the OP's macro by one or more PROC FREQ calls.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
@FreelanceReinhard wrote:
@data_null__: Yes, I assume it's the same CI (but haven't checked). However, I guess it would not help much to replace the data step formula in the OP's macro by one or more PROC FREQ calls.
I'm not suggesting that you loop on PROC FREQ I was asking if the entire thing can be done with PROC FREQ.
It might be that looping on PROC FREQ is better than "this" program.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
@data_null__ wrote:
... I was asking if the entire thing can be done with PROC FREQ.
I don't think so. PROC FREQ offers a variety of plots, but the type of graph produced by the OP's program is not among these, I think. Plotting coverage probabilities is rather something for researchers in mathematical statistics than for the target audience of PROC FREQ, i.e. users who are analyzing data.
Learn how use the CAT functions in SAS to join values from multiple variables into a single value.
Find more tutorials on the SAS Users YouTube channel.
SAS Training: Just a Click Away
Ready to level-up your skills? Choose your own adventure.