- Home
- /
- Programming
- /
- Programming
- /
- How to compare current observation with next observation?
- RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Hello,
I want to compare some variables in both observations nearby each time in the transposed dataset. If any variable within name, age, term and rate is different, then add a new column flag on first observation.
eg. Observation1: subject=301001, folderseq=1 recordposition=1 _NAME_=new_val;
Observation2: subject=301001, folderseq=1, recordposition=1 and _NAME_=newval; because the variable age is different. so new variable flag='Modified' will be added onto the record with _NAME_=new_val . Could you please help me to look this? Thank you in advance!
Best Regards
Leo
data test1;
input subject & $8. folderseq RecordPosition _NAME_ & $10. name & $12. age & $30. term & $48. rate & $30.;
cards;
301001 1.0 1 new_val Frank 21 headache 6
301001 1.0 1 newval "" 21|Modified|20 headache|Modified|Headache 6|Modified|5
301001 1.0 2 new_val Frank 20 pain 5
301001 1.0 2 newval "" "" "" ""
301002 2.0 1 new_val Annie 31 abcd 4
301002 2.0 1 newval "" "" abcd|Modified|abc ""
301003 4.0 0 new_val Andy 40 b1 3
301003 4.0 0 newval Andy|added 40|added b1|added 3|added
;
run;
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Please provide data in usable form.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
data WORK.TEST2;
infile datalines dsd truncover;
input subject:$8. folderseq:32. RecordPosition:32. _NAME_:$10. name:$12. age:$30. term:$48. rate:$30.;
datalines;
301001 1 1 new_val Frank 21 headache 6
301001 1 1 newval "" 21|Modified|20 headache|Modified|Headache 6|Modified|5
301001 1 2 new_val Frank 20 pain 5
301001 1 2 newval "" "" "" ""
301002 2 1 new_val Annie 31 abcd 4
301002 2 1 newval "" "" abcd|Modified|abc ""
301003 4 0 new_val Andy 40 b1 3
301003 4 0 newval Andy|added 40|added b1|added 3|added
;;;;
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Sorry, but there are several problem with the step you have posted, please fix them. Also you want to add the expected result for the data provided.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Sorry Andreas! Please find the enclosed updated code below. Thanks
data work.test2 ;
infile datalines dsd dlm='|' truncover;
input subject :$8. folderseq RecordPosition _NAME_ :$10. name :$12.
age :$30. term :$48. rate :$30.
;
datalines4;
301001|1|1|new_val|Frank|21|headache|6
301001|1|1|newval|""""""|"21|Modified|20"|"headache|Modified|Headache"|"6|Modified|5"
301001|1|2|new_val|Frank|20|pain|5
301001|1|2|newval|""""""|""""""|""""""|""""""
301002|2|1|new_val|Annie|31|abcd|4
301002|2|1|newval|""""""|""""""|"abcd|Modified|abc"|""""""
301003|4|0|new_val|Andy|40|b1|3
301003|4|0|newval|"Andy|added"|"40|added"|"b1|added"|"3|added"
;;;;
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Could you please help me to look this again? any suggestions would be appreciated.
Thanks
Leo
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
All of the data you provided is different. So as you described the rule the new flag should be true for EVERY observation.
Can you provide some data that can actually be used the test whether the code can tell the difference between values that change and those that don't?
Also make sure to provide the answer key.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Hello, Thank you. I have updated this data. I hope to add a new variable flag=M or flag=A to previous observation when there is delimiter('|') and word(Modified or added) at current observation.
data work.data_1 ;
infile datalines dsd dlm='|' truncover;
input subjectid :$8. folderseq RecordPosition subjectname :$12.
age :$15. term :$48. rate :$15.
;
datalines4;
301001|1|1|Frank|20|headache|6
301001|1|1|Frank|20|"headache|Modified|Headache"|6
301001|1|2|Frank|20|Stomachache|5
301001|1|2|Frank|20|Stomachache|5
301001|1|3|Frank|20|Insomnia|5
301001|1|3|"Frank|added"|"20|added"|"Insomnia|added"|"5|added"
301002|2|1|Annie|31|fever|4
301002|2|1|Annie|31|fever|"4|Modified|5"
301002|2|2|Annie|31|Cough|4
301002|2|2|Annie|31|Cough|4
301004|4|0|Andy|40|b1|3
301004|4|0|"Andy|added"|"40|added"|"b1|added"|"3|added"
;;;;
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
What are the key variables here? How do you want to determine which observations should be compared?
If is much easier to compare the current observation's values to the previous observations value than the next value.
It is much easier for the data step to remember what it has seen than to predict the future.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Thank you for your help! you are right. the key variables are separately subject, folderseq and recordposition.
the other variables (name, age, term and rate )will be compared at two different datasets.
actually, I need to review routinely datasets current and previous datasets. The frequency is each month. To find out any update record, added record and deleted record from current dataset after comparing with previous dataset. I have written some code. but I do not know how to put a flag to current dataset to display updated record/new record/removed record.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Are you trying to do something like this?
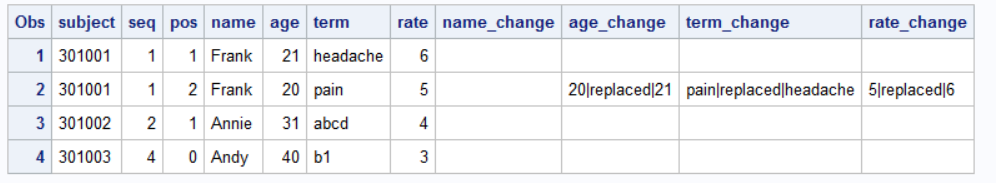
So you have some data that is keyed by SUBJECT SEQ POS.
data have ;
input subject $ seq pos name $ age term $ rate ;
cards;
301001 1 1 Frank 21 headache 6
301001 1 2 Frank 20 pain 5
301002 2 1 Annie 31 abcd 4
301003 4 0 Andy 40 b1 3
;And you want to check if there are changes in NAME AGE TERM or RATE.
So you might want to make a check like this:
%macro check(var);
_name_="&var";
change=' ';
if &var ne lag_&var then do;
if missing(lag_&var) then change = catx('|',&var,'added');
else if missing(&var) then change = catx('|','removed',lag_&var);
else change=catx('|',&var,'replaced',lag_&var);
end;
output;
%mend check;
data diffs ;
set have;
by subject seq pos ;
lag_name=lag(name);
lag_age=lag(age);
lag_term=lag(term);
lag_rate=lag(rate);
if not first.seq ;
length _name_ $32 change $100 ;
%check(name)
%check(age)
%check(term)
%check(rate)
keep subject seq pos _name_ change ;
run;Which you can then transpose
proc transpose data=diffs out=wide(drop=_name_) suffix=_change;
by subject seq pos;
id _name_;
var change;
run;And merge with the original data
data want;
merge have wide;
by subject seq pos;
run;
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Yes. I did. but it does not work. how to put this flag to corresponding records(updated, added and deleted) onto this NEW's dataset? Could you please help me to look this? Thank you in advance!
data work.OLD ;
infile datalines dsd dlm='|' truncover;
input name :$12. age subject :$8. folderseq RecordPosition term :$48.
rate
;
datalines4;
Frank|20|301001|1|1|Headache|5
Frank|20|301001|1|2|pain|5
Annie|31|301002|2|1|abc|4
Annie|31|301002|2|2|abcd|4
Jessica|38|301005|5|0|a1|3
;;;;
data work.NEW ;
infile datalines dsd dlm='|' truncover;
input name :$12. age subject :$8. folderseq RecordPosition term :$48.
rate
;
datalines4;
Frank|21|301001|1|1|headache|6
Frank|20|301001|1|2|pain|5
Annie|31|301002|2|1|abcd|4
Annie|31|301002|2|2|abcd|4
Andy|40|301004|4|0|b1|3
;;;;
data have1;
set OLD;
run;
data have2;
set NEW;
run;
%let key_col=subject folderseq RecordPosition;
/* %let compare_col=name age sex rate; */
%let key_col_cnt=%sysfunc(countw(&key_col));
/* Code to find compare column list - START */
proc contents data=have2 out=contents2 noprint;
run;
%macro where_cond;
%do i=1 %to &key_col_cnt;
%if &i < &key_col_cnt %then
%do;
%let col=%sysfunc(scan( &key_col,&i));
"&col" ","
%end;
%else
%do;
%let col=%sysfunc(scan( &key_col,&i));
"&col" ", "
%end;
%end;
%mend;
proc sql noprint;
select name into :compare_col separated by ' '
from contents2
where name not in (%where_cond);
quit;
%put COMPARE COLUMN = &compare_col ;
/* Code to find compare column list - END */
%macro select_col;
%do i=1 %to &key_col_cnt;
%let col=%sysfunc(scan( &key_col,&i));
coalesce(a.&col,b.&col) as &col,
%end;
%mend;
%macro join_cond;
%do i=1 %to &key_col_cnt;
%let col=%sysfunc(scan( &key_col,&i));
a.&col =b.&col and
%end;
%mend;
proc sort data=have1 out=have1_sorted;
by &key_col ;run;
proc sort data=have2 out=have2_sorted;;
by &key_col ;run;
proc transpose data=have1 out=have1_trans (rename= (_name_=col_name));
by &key_col ;
var &compare_col ;
run;
proc transpose data=have2 out=have2_trans(rename= (_name_=col_name)) ;
by &key_col ;
var &compare_col;
run;
proc sql;
create table compare_table
as
select %select_col
coalesce(a.col_name,b.col_name) as variable_name
,strip(a.col1) as old_val
,strip(b.col1) as new_val
,case when strip(a.col1) <> strip(b.col1) and a.subject= b.subject and a.folderseq =b.folderseq then 'Modified'
when strip(a.col1) <> strip(b.col1) and (a.subject='' or a.folderseq=.) then 'Added'
when strip(a.col1) <> strip(b.col1) and (b.subject='' or b.folderseq=.) then 'Deleted' end as remark
from have1_trans a
full join have2_trans b
on %join_cond
a.col_name = b.col_name;
quit;
Data compare_table_2;
set compare_table;
if remark in ("Added") then newval=strip(strip(new_val)||"|"||"Added");
else if remark in ("Deleted") then newval=catx('|',"@","Deleted",old_val);
else if remark in ("Modified") then newval=catx('|',new_val,"Modified",old_val);
run;
/* Transpose second time */
proc transpose data=compare_table_2 out=have2_new_trans_2;
by subject FolderSeq RecordPosition;
var new_val newval;
id variable_name;
run;
data temp1;
set have2_new_trans_2;
delim='|';
if scan(age,2,delim)="Modified" then flag='M';
else if scan(age,2,delim)="Added" then flag='A';
else if scan(age,2,delim)="Deleted" then flag='D';
run;
Learn how use the CAT functions in SAS to join values from multiple variables into a single value.
Find more tutorials on the SAS Users YouTube channel.
SAS Training: Just a Click Away
Ready to level-up your skills? Choose your own adventure.