- RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
I want to calculate the MD which represents the class separation between these two classes.(variable intercepts are shown in the picture attached) .
The intercepts for the first group are 0.35,-0.2,0.6,-0.75,0.35,-0.2,0.6,-0.75,0.35,-0.2
The intercepts for the second group are 0,0,0,0,0,0,0,0,0,0
How to calculate the MD beween these two latent classes(in mixture modeling)? I don't know how to edit the appropriate syntax. Thanks in advance!

Accepted Solutions
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
The Euclidean distance formula you are using is the distance between two 10 -dimensional points. The general ED formula for the distance between points p and q is
ED = sqrt( (p1-q1)**2 + (p2-q2)**2 + ... + (p10 -q10)**2 )
In this case p is the set of 10 X values and q is the center (0,0,0,...0).
So you need to produce a nonsingular 10x10 covariance matrix if you want to compute the Mahalanobis distance.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
My original syntax as follows:
/* simplest approach: x and center are row vectors */
start mahal1(x, center, cov);
y = (x - center)`; /* col vector */
d2 = y` * inv(cov) * y; /* explicit inverse. Not optimal */
return (sqrt(d2));
finish;
x = {0.35 0,
-0.2 0,
0.6 0,
-0.75 0,
0.35 0,
-0.2 0,
0.6 0,
-0.75 0,
0.35 0,
-0.2 0}; /* test it */
center = {0.015 0};
cov = {0.1225 -0.07 0.21 -0.2625 0.1225 -0.07 0.21 -0.2625 0.1225 -0.07,
-0.07 0.04 -0.12 0.15 -0.07 0.04 -0.12 0.15 -0.07 0.04,
0.21 -0.12 0.36 -0.45 0.21 -0.12 0.36 -0.45 0.21 -0.12,
-0.2625 0.15 -0.45 0.5625 -0.2625 0.15 -0.45 0.5625 -0.2625 0.15,
0.1225 -0.07 0.21 -0.2625 0.1225 -0.07 0.21 -0.2625 0.1225 -0.07,
-0.07 0.04 -0.12 0.15 -0.07 0.04 -0.12 0.15 -0.07 0.04,
0.21 -0.12 0.36 -0.45 0.21 -0.12 0.36 -0.45 0.21 -0.12,
-0.2625 0.15 -0.45 0.5625 -0.2625 0.15 -0.45 0.5625 -0.2625 0.15,
0.1225 -0.07 0.21 -0.2625 0.1225 -0.07 0.21 -0.2625 0.1225 -0.07,
-0.07 0.04 -0.12 0.15 -0.07 0.04 -0.12 0.15 -0.07 0.04};
md1 = mahal1(x, center, cov);
print md1;
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Here is the output, the program presented error messages:
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
I would recommend that you move your question to the IML forum here:
https://communities.sas.com/t5/SAS-IML-Software-and-Matrix/bd-p/sas_iml
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
As of SAS/IML 12.1, which shipped in August 2012 as part of SAS 9.3m2, the MAHALANOBIS function is distributed as part of SAS/IML software. You do not need to define your own function.
The doc has an example. For your example, you list a 10x10 covariance matrix. That means that the CENTER vector should be a 1x10 row matrix and the X matrix should have 10 columns, not 10 rows. Also, the covariance matrix must be nonsingular. Yours appears to be singluar.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Thanks, Dr. Wicklin! I have read the article concerning the calculation of MD within SAS before, but I am so sorry that I don't truly understand what does the "center" mean in SAS.
In your blog, the article says"Given an observation x from a multivariate normal distribution with mean μ and covariance matrix Σ, the squared Mahalanobis distance from x to μ is given by the formula d2 = (x - μ)T Σ -1 (x - μ). You can use this definition to define a function that returns the Mahalanobis distance for a row vector x, given a center vector (usually μ or an estimate of μ) and a covariance matrix:"
In my word, the center vector in my example is the 10 variable intercepts of the second class, namely 0,0,0,0,0,0,0,0,0,0.
I know I am wrong, but don't know what is wrong. I hope that you can help me with my problem. Thank you!
proc iml;
/* simplest approach: x and center are row vectors */
start mahal1(x, center, cov);
y = (x - center)`; /* col vector */
d2 = y` * inv(cov) * y; /* explicit inverse. Not optimal */
return (sqrt(d2));
finish;
x = {0.35 -0.2 0.6 -0.75 0.35 -0.2 0.6 -0.75 0.35 -0.2,
0 0 0 0 0 0 0 0 0 0}; /* test it */
center = {0 0 0 0 0 0 0 0 0 0 0};
cov = {0.1225 -0.07 0.21 -0.2625 0.1225 -0.07 0.21 -0.2625 0.1225 -0.07,
-0.07 0.04 -0.12 0.15 -0.07 0.04 -0.12 0.15 -0.07 0.04,
0.21 -0.12 0.36 -0.45 0.21 -0.12 0.36 -0.45 0.21 -0.12,
-0.2625 0.15 -0.45 0.5625 -0.2625 0.15 -0.45 0.5625 -0.2625 0.15,
0.1225 -0.07 0.21 -0.2625 0.1225 -0.07 0.21 -0.2625 0.1225 -0.07,
-0.07 0.04 -0.12 0.15 -0.07 0.04 -0.12 0.15 -0.07 0.04,
0.21 -0.12 0.36 -0.45 0.21 -0.12 0.36 -0.45 0.21 -0.12,
-0.2625 0.15 -0.45 0.5625 -0.2625 0.15 -0.45 0.5625 -0.2625 0.15,
0.1225 -0.07 0.21 -0.2625 0.1225 -0.07 0.21 -0.2625 0.1225 -0.07,
-0.07 0.04 -0.12 0.15 -0.07 0.04 -0.12 0.15 -0.07 0.04};
md1 = mahal1(x, center, cov);
print md1;
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
The 'center' matrix is an estimate of the center of the population. for example, the center is often taken to the be the sample mean of the data, just as the covariance estimate is the sample covariance.
In the code that you posted, there are two things wrong:
1. The Center vector has 11 elements, not 10.
2. The covariance matrix is singular. The covariance must be nonsingular to compute the Mahalanobis distance.
For demonstration purposes, the following code adds a multiple of the identity matrix to your covariance. In real life, you'll have to figure out how to get a nonsingular matrix. Notice that I call the built-in MAHALANOBIS function:
proc iml;
x = {0.35 -0.2 0.6 -0.75 0.35 -0.2 0.6 -0.75 0.35 -0.2};
center = {0 0 0 0 0 0 0 0 0 0};
cov = {0.1225 -0.07 0.21 -0.2625 0.1225 -0.07 0.21 -0.2625 0.1225 -0.07,
-0.07 0.04 -0.12 0.15 -0.07 0.04 -0.12 0.15 -0.07 0.04,
0.21 -0.12 0.36 -0.45 0.21 -0.12 0.36 -0.45 0.21 -0.12,
-0.2625 0.15 -0.45 0.5625 -0.2625 0.15 -0.45 0.5625 -0.2625 0.15,
0.1225 -0.07 0.21 -0.2625 0.1225 -0.07 0.21 -0.2625 0.1225 -0.07,
-0.07 0.04 -0.12 0.15 -0.07 0.04 -0.12 0.15 -0.07 0.04,
0.21 -0.12 0.36 -0.45 0.21 -0.12 0.36 -0.45 0.21 -0.12,
-0.2625 0.15 -0.45 0.5625 -0.2625 0.15 -0.45 0.5625 -0.2625 0.15,
0.1225 -0.07 0.21 -0.2625 0.1225 -0.07 0.21 -0.2625 0.1225 -0.07,
-0.07 0.04 -0.12 0.15 -0.07 0.04 -0.12 0.15 -0.07 0.04};
cov = cov + I(10); /* HACK: make the cov matrix nonsingular for demonstration */
md1 = mahalanobis(x, center, cov);
print md1;
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Hello again! With the default MD function and the previous syntax, the MD value is 0.83. But in the simulation study, the author fixed the MD value at 1.5 for small separation between the 2 classes. (As you can see in the 2 pictures attached) Thank you for your light on this difference!
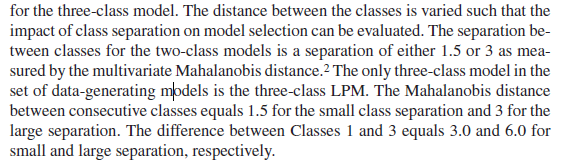
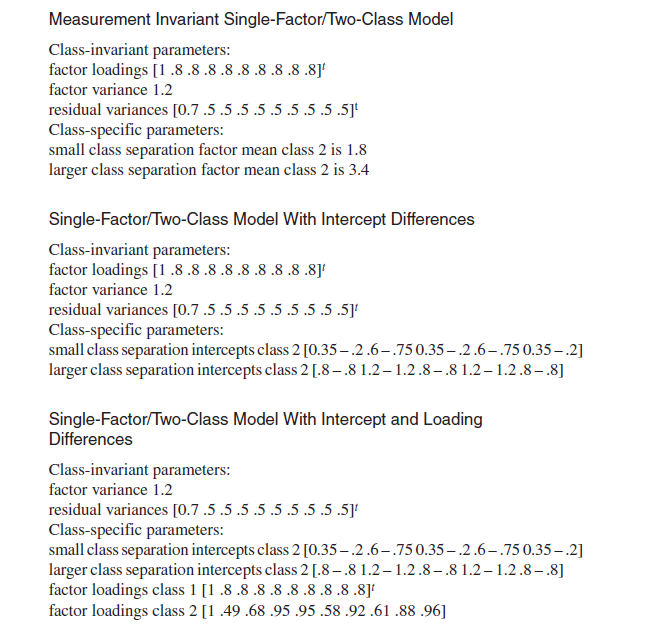
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Please READ my response which says
"For demonstration purposes, the following code adds a multiple of the identity matrix to your covariance. In real life, you'll have to figure out how to get a nonsingular matrix."
Also the program says
/* HACK: make the cov matrix nonsingular for demonstration */In other words, I am not claiming that the MD is 0.83. You have provided a singular matrix, which means that the MD is undefined. I modified your covariance matrix in order to show you how to calculate the MD. You won't match someone else's results unless you use the same inputs (mean vector and covariance matrix) that they used.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Yes, the covariance matrix in fact should be a 2*2 matrix because the sample has 2 dimensions(2 groups of observed variable intercepts).
cov = {0.2589 0,
0 0};
The x should be
x = {0.35 -0.2 0.6 -0.75 0.35 -0.2 0.6 -0.75 0.35 -0.2};
But the center. You said the center is the sample mean of the data.
The center should be {0.015,0};
But this syntax still did not work. I am a green hand in statistics. Thank you for your help!
proc iml;
x = {0.35 -0.2 0.6 -0.75 0.35 -0.2 0.6 -0.75 0.35 -0.2};
center = {0.015 0};
cov = {0.2589 0,
0 0};
md1 = mahalanobis(x, center, cov);
print md1;
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Let p be the number of variables in your problem.
Then
- X should be a matrix that has p columns.
- 'center' should be a 1 x p row vector.
- the covariance matrix should be a symmetric nonsingular p x p matrix.
In your problem, do you have 2 variables or 10?
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Sorry to repeat myself, but you still do not have the dimensions correct.
1. If the mean is 1x2 and the covariance matrix is 2x2, then the X matrix must have two columns.
2. The covariance matrix must be nonsingular. Your matrix cov = {0.2589 0, 0 0} is singular.
3. If you expect to get one "distance" (you say the answer is 1.5), then X would have 1 row. If you specify 10 rows, you are saying that there are 10 points and you want to know the distance from each point to the center.
If you show the Euclidean calculation (for which you claim you get the correct answer) that might help us to make sense of what you are trying to do.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
The Euclidean distance formula you are using is the distance between two 10 -dimensional points. The general ED formula for the distance between points p and q is
ED = sqrt( (p1-q1)**2 + (p2-q2)**2 + ... + (p10 -q10)**2 )
In this case p is the set of 10 X values and q is the center (0,0,0,...0).
So you need to produce a nonsingular 10x10 covariance matrix if you want to compute the Mahalanobis distance.
April 27 – 30 | Gaylord Texan | Grapevine, Texas
Registration is open
Walk in ready to learn. Walk out ready to deliver. This is the data and AI conference you can't afford to miss.
Register now and lock in 2025 pricing—just $495!
- Ask the Expert: Wie kann ich Visual Studio Code als SAS Entwicklungsumgebung nutzen? | 11-Dec-2025
- DCSUG Online Winter Webinar | 11-Dec-2025
- Ask the Expert: Marketing Decisions Excellence: Turning Insights Into Business Impact | 16-Dec-2025
- SAS Bowl LVII, SAS Data Maker and Synthetic Data | 17-Dec-2025
- SAS Innovate 2026 | Grapevine, Texas | 27-Apr-2026


