- Home
- /
- SAS Communities Library
- /
- Data Library Comparison Macro %COMPARE_ALL
- RSS Feed
- Mark as New
- Mark as Read
- Bookmark
- Subscribe
- Printer Friendly Page
- Report Inappropriate Content
Data Library Comparison Macro %COMPARE_ALL
- Article History
- RSS Feed
- Mark as New
- Mark as Read
- Bookmark
- Subscribe
- Printer Friendly Page
- Report Inappropriate Content
This was a paper that was supposed to be presented at PharmaSUG 2020 this year, but due to the cancellation of the conference I still wanted to create a page with the paper and macro for download to share with everyone. This is a macro that has been extremely convenient whenever I need to compare frozen libraries of data together to get a report of what has changed. It removes the tediousness of having to compare individual datasets and makes the output easier to read.
Abstract
Reproducible research and sharing of data with repositories are becoming more standard, and so the freezing of data for specific analyses is more crucial than ever before. Maintaining multiple data freezes requires knowing what changed within the data from one version to another. In SAS there is the COMPARE procedure that allows the user to compare two data sets to see potential new variables, lost variables, and changes in values. Relying on the COMPARE procedure can be tedious and cumbersome when maintaining a database containing several data sets. The COMPARE_ALL macro was written to ease this burden by generating a Microsoft Excel report of a comparison of two data libraries instead of just two data sets. The report indicates any new or lost data sets, variables or observations and checks for changed data values within all variables. Multiple ID variables can be specified and the macro will determine which variables are relevant with each data set for comparison. The COMPARE_ALL macro is a fantastic tool for managing multiple versions of the same SAS database.
Introduction
The research field of clinical oncology has many reasons for keeping copies, or freezes, of study data including: standardized reporting such as to the Data Safety Monitoring Board (DSMB), sharing data to a repository such as Project Data Sphere, or when publishing the analysis in an abstract or manuscript. Studies typically have multiple data freezes over several years, and keeping a log of the changes is difficult. The COMPARE Procedure is useful for comparing one data set at a time, but studies often contain many data sets and using the COMPARE Procedure on each one is tedious. The COMPARE_ALL macro was created to make an easy to read report in an Excel that compares entire SAS libraries to check for various changes such as new or lost data sets, new or lost variables, change to variable attributes, and change to variable values. The generated report is a powerful tool for summarizing data changes and ensuring that the data changes are expected.
Report Features
The COMPARE_ALL macro produces an Excel report with four different types of tables. The first is an overall summary of the data sets within the base and comparison libraries, the second is a summary of the variables within a specific data set as well as several variable attributes, the third is a summary of the types of data changes within each data set, and the last is a variable level change summary.
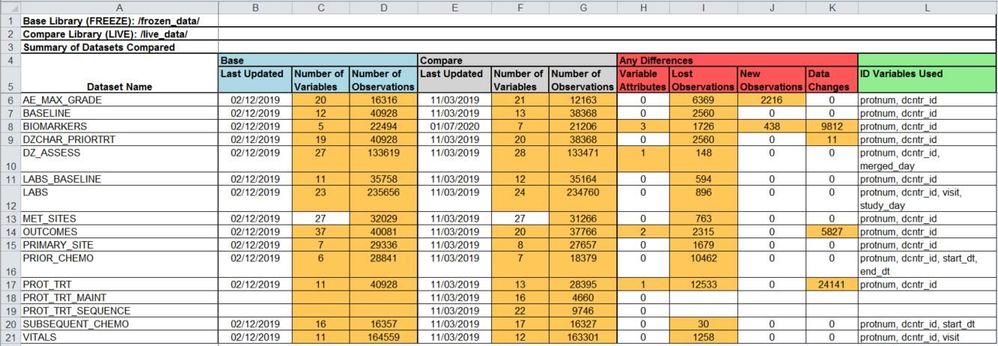
Table Type 1: Library Summary
This table is only listed once in the report and is always listed first. The table lists each data set in either the base or comparison library in alphabetical order. If a data set exists within both libraries several comparisons are highlighted:
- Date data sets were last updated
- Number of variables
- Number of observations
- Number of variable attributes that changed (e.g. length, type, label)
- Number of lost observations (when an ID variable is specified)
- Number of new observations (when an ID variable is specified)
- Number of data changes (variable value changes for same observation/ID combination)
The table also indicates which ID variables in the ID list are available to be used in comparisons for the given data set. The ID variables used can be different within each data set.
The report automatically uses colors to highlight data changes and to reference the same items across tables. Attributes for the base library will always be highlighted blue, the compare library will be grey, ID variables will be green, and data differences will be red. In table 1 any changes between base and compare will be highlighted orange to draw the user’s attention immediately. The header displays the file paths of both the base and comparison libraries so that the user can ensure the correct libraries are being compared.
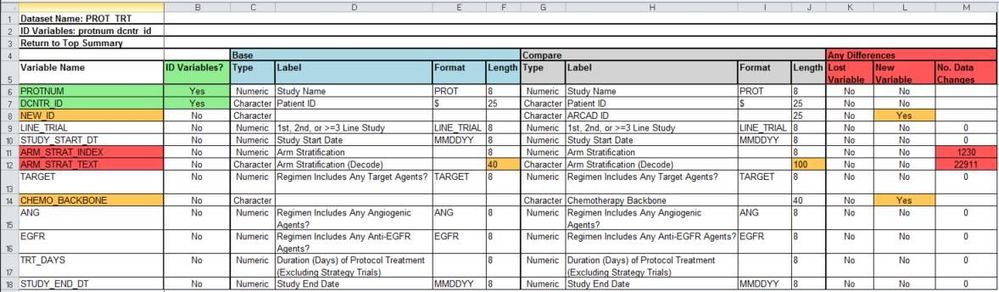
Table Type 2: Data Set Level Summary
The macro report will include a data set level summary for each data set that exists within both the base and compare libraries. Variables are listed in the same order as they appear in the data set by default, but can be listed alphabetically as well. Each variable indicates if it is an ID variable, has the TYPE, LABEL, FORMAT, and LENGTH attributes listed as well as whether the variable is lost (in base but not in compare), new (in compare but not in base), and for how many observations did the variable’s values change for a given ID combination.
Variable names or attributes are highlighted in three cases: if they are an ID variable (green), if an attribute such as typing has changed (orange) and if any of the “Any Differences” columns are not equal to “No” (orange) or if the number of data changes are greater than zero (red). This draws the user’s attention to the variables and attributes that are of interest for the difference report. The header lists the data set name and gives a list of the ID variables used for that particular data set.
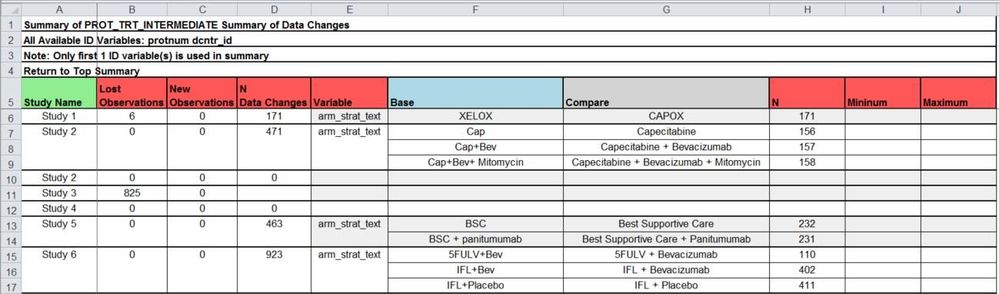
Table Type 3: Data changes summary
The third table type is optional and displays a summary of the data changes either within a data set grouping by either the first N (specified by IDSUMTABLE parameter) ID variable(s) available or across all observations. The summary is useful to see a quick description of all the changes, whether all of the changes are coming from the same study or patient, or if the changes are spread out. This also gives a chance to see if related variables are also changing. For example being able to immediately check if the number of patients having death dates has increased the same amount as the number of patients having a survival status changed from alive to dead.
The above figure summarizes all of the variable changes within the first ID variable as well as displaying the number of lost observations, new observations, and data changes. For a meta-study such as the one in this figure, this is valuable to see if the changes are coming from the expected studies or if a programming error could have caused the changes. The summary for each variable changes depending on criteria:
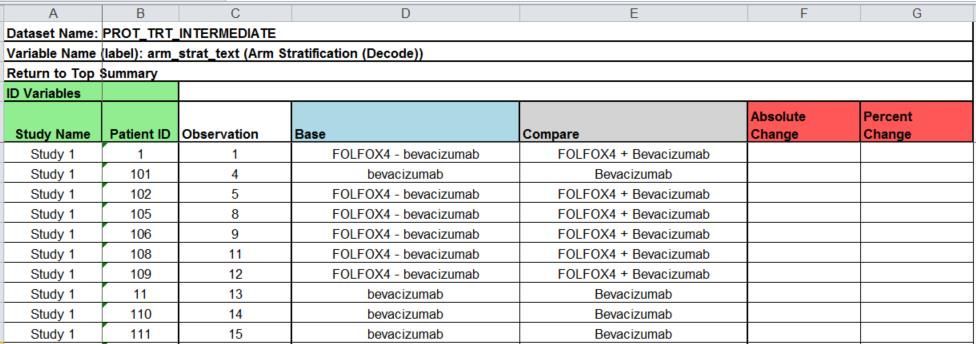
- Comparing two character variables: displays a cross tab of the before and after values with frequencies (see arm_strat_text)
- Comparing two numeric variables having less than or equal to macro parameter CROSSTAB_THRESHOLD’s unique combinations: displays a cross tab of the before and after values with frequencies
- Comparing two numeric variables having greater than the macro parameter CROSSTAB_THRESHOLD’s unique combinations: displays the number of times values changed from missing to non-missing, non-missing to missing, and non-missing to non-missing. There will be a frequency and minimum and maximum change values where appropriate.
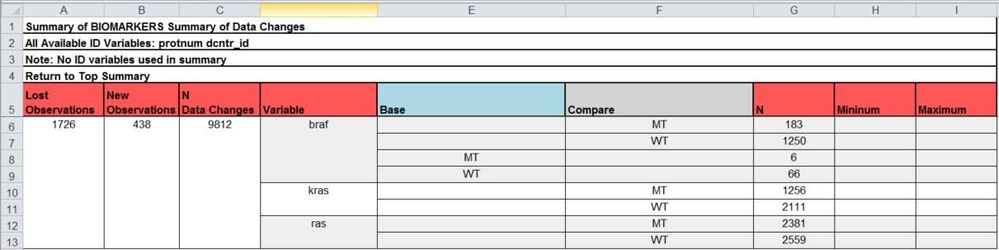
The header of the table indicates the data set name and ID variable used to be clear to the user. It is also possible to specify IDSUMTABLE=0 and summarize across all observations:
The above figure summarizes all of the variable changes, lost observations, new observations, and data changes across all observations of the data sets. This is a quick way to summarize all variable level changes when there are many changes across numerous ID variable levels.
Table Type 4: Variable Change Summary
The fourth table summary is similar to the output given by the COMPARE procedure. Each variable that has data changes will have its own worksheet to show individual observation changes.
The listing will include each ID variable, observation number in the data set, the base data set value, the compare data set value, and the absolute and percent changes if both variables are numeric and non-missing. The header of the table lists the data set, variable name and label. The listing is straightforward and mimics the output of the COMPARE procedure.
Navigating the Report
The report has the potential to create a large amount of worksheets which can make it difficult to navigate quickly to the desired summary. The COMPARE_ALL macro accounts for this by inserting hyperlinks into each table to allow the user to jump to the appropriate summary and back. Examples of the hyperlinks are:
- Within the Type 1 worksheet the user can click the name of any data set to jump to that data set’s Type 2 data set summary page.
- Within each Type 2 data set summary page any variable that is marked red for data changes can be clicked on to jump to that variable’s type 4 variable listing.
- Each Type 2, 3, and 4 worksheet includes a link in the header to either jump back to the Type 1 worksheet or to drill back up a level (e.g. Type 4 listing back to the Type 2 data set listing)
These hyperlinks allow the user to navigate the report much more efficiently.
Calling the COMPARE_ALL Macro
The COMPARE_ALL macro itself is straightforward available due to the small amount of parameters needed. Using the macro requires access to ODS EXCEL within SAS. There are three required parameters and four optional parameters:
- Required parameters:
- BASE: Library name to be considered the old data for comparisons. Note that the libname must already be specified prior to the macro.
- COMPARE: Library name to be considered the new data for comparisons. Note that the libname must already be specified prior to the macro.
- OUTDOC: The destination filepath and filename of the XLSX file that will contain the report
- Optional parameters:
- ID: A space delimited list of variable names to be used as ID variables. Note that not all data sets in the same library have to contain all of the ID variables. The macro will match any available ID variable in the list to a given data set in the order they are listed. ID variables not in a given data set are ignored for that comparison.
- IDSUMTABLE: Determines how many of the available ID variables are used to produce the Table 3 Summary
- SELECT: Allows the user to specify which data sets in the library are included in the report. The user can specify a space delimited list of the desired data sets. If this option is used then a message is shown in the Type 1 worksheet to indicate that not all of the data sets in the libraries are being shown. The data sets in the SELECT statement must exist in both the BASE and COMPARE libraries.
- CROSSTAB_THRESHOLD: Determines the threshold for the number of unique variable value changes before summarizing the changes as non-missing and missing to save vertical space. The value defaults to 15 and must be greater than or equal to 1.
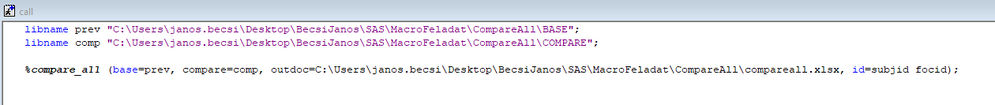
The following is the macro call that leads to the images in figure 1 to figure 4 (file paths and study names have been masked for confidentiality):
libname live '/live_data/';
libname freeze '/frozen_data/';
options fmtsearch=(live work library);
%compare_all(
base=freeze,
compare=live,
id=protnum dcntr_id merged_day visit study_day start_dt end_dt,
outdoc=database_changes.xlsx,
select=,
idsumtable=1);The libraries are predefined in LIBNAME statements. The FMTSEARCH option is enabled in order to activate the formats of the library in order to make the values in the comparisons make sense. The most complicated parameter is the ID option. There are seven ID variables listed, but a majority of the data sets will only use the first two listed ID variables due to not having the other five. There are several data sets in these libraries where there are multiple rows per patient and times such as MERGED_DAY and START_DT are necessary to compare the appropriate observations. The macro will search for all seven listed ID variables in each data set and subset down to only the variables that exist in both the base and compare data sets.
Conclusion
The COMPARE_ALL macro is a powerful tool to efficiently compare two SAS libraries versus running multiple COMPARE Procedure calls for each data set in the libraries. The generated report is clean and straightforward to read with built in navigation for ease of use. The COMPARE_ALL macro will be useful for any programmer working with clinical trial data and is available to be shared.
Contact Information
I can be contacted for questions or issues with using the macro. I'll be updating this page with the final paper and presentation when they are available. I normally program in Linux so there is the potential for issues in Windows SAS that I do not normally encounter.
Name: Jeffrey Meyers
Enterprise: Mayo Clinic
- Mark as Read
- Mark as New
- Bookmark
- Permalink
- Report Inappropriate Content
Thank you @JeffMeyers for sharing your work!
- Mark as Read
- Mark as New
- Bookmark
- Permalink
- Report Inappropriate Content
Very useful macro. Thank you.
- Mark as Read
- Mark as New
- Bookmark
- Permalink
- Report Inappropriate Content
Thank your @JeffMeyers very useful macro. I have couple of questions.
1. I would like to change the color of the text where it creates the hyperlinks for example in " Top summary" sheet. in columns with with dataset name will hyperlink to other sheets in the file. I want to change the color of dataset to blue so we know its as hyperlink. I played around the first proc report column styles but it did not worked for me. Any ideas
2. Is this macro through any notes/ warning to indicate if one of the library missing the dataset lets say one have 3 datasets and another have 2? . I understand that 'select ' macro variable have the ability to select the datasets.
3. Is it gives any indication or notes or warning about if 'compare' dataset missing the one of the variable in the ID macro variable compare to base dataset? Ex: lets say base have variables 'usubjid studyid siteid' but compare have ' usubjid studyid' only. If we call ID macro variable with 'usubjid studyid siteid', is it gives any notes or warning one of the dataset missing the values or it will just run with the common variables?
- Mark as Read
- Mark as New
- Bookmark
- Permalink
- Report Inappropriate Content
Thank you @JeffMeyers for your macro, it's really powful.
I have one suggestion, if user use UTF-8 SAS, he need change "A0"x to 'C2A0'x in PROC REPORT, otherwise SAS will occur "ERROR: Invalid characters were present in the data."
in my side, I just made some modification of your code .
%local nbsp;
data _null_;
length nbsp $10;
if "%SYSFUNC(GETOPTION(encoding))" = "%str(WLATIN1)" then nbsp='A0'x;
else if "%SYSFUNC(GETOPTION(encoding))" = "%str(UTF-8)" then nbsp='C2A0'x;
call symput('nbsp',put(nbsp,$hex8.));
run;
define n6 / across " "x style={borderbottomstyle=none borderbottomcolor=white};
define n5 / across " "x style={borderbottomstyle=none borderbottomcolor=white background=lightgreen};
- Mark as Read
- Mark as New
- Bookmark
- Permalink
- Report Inappropriate Content
Hello @SASuserlot
My apologies for not getting back to you sooner.
1. I would like to change the color of the text where it creates the hyperlinks for example in " Top summary" sheet. in columns with with dataset name will hyperlink to other sheets in the file. I want to change the color of dataset to blue so we know its as hyperlink. I played around the first proc report column styles but it did not worked for me. Any ideas
--You can do this within a compute block. There should be a number of CALL DEFINE statements that are assigning the URL for the hyperlinks. If you go to where these are made and at the same time add something like CALL DEFINE('column name','style/merge','style={color=blue}'); it should color the font blue. BACKGROUNDCOLOR will change the fill color.
2. Is this macro through any notes/ warning to indicate if one of the library missing the dataset lets say one have 3 datasets and another have 2? . I understand that 'select ' macro variable have the ability to select the datasets.
--If a dataset is in one library but not the other it would only be visible in the first sheet summary. There should be dates for the dataset that exists and nothing for the one that doesn't
3. Is it gives any indication or notes or warning about if 'compare' dataset missing the one of the variable in the ID macro variable compare to base dataset? Ex: lets say base have variables 'usubjid studyid siteid' but compare have ' usubjid studyid' only. If we call ID macro variable with 'usubjid studyid siteid', is it gives any notes or warning one of the dataset missing the values or it will just run with the common variables?
--There are no warnings or notes to indicate which ID variables exist or don't in each dataset. These are only listed on the first page summary. This is because each dataset can have a different combination of variables so there was no reason to add warnings in this case.
- Mark as Read
- Mark as New
- Bookmark
- Permalink
- Report Inappropriate Content
Hello @NovGetRight . Thank you for the suggestion. I have come across situations like this as well and am learning to account for them in future programs.
- Mark as Read
- Mark as New
- Bookmark
- Permalink
- Report Inappropriate Content
Thank you @JeffMeyers for the macro.
Have an error occured and cannot figure out what is the problem:
I have downloaded the file and modified only the %local nbsp part suggested above. Can you please check it.
Thanks in advance,
Janos
- Mark as Read
- Mark as New
- Bookmark
- Permalink
- Report Inappropriate Content
@JanosBecsi since the macro uses PROC and DATA steps, you cannot call it within PROC SQL (which you seem to have done).
- Mark as Read
- Mark as New
- Bookmark
- Permalink
- Report Inappropriate Content
- Mark as Read
- Mark as New
- Bookmark
- Permalink
- Report Inappropriate Content
Then why does your log show
else . end,immediately after the macro call?
- Mark as Read
- Mark as New
- Bookmark
- Permalink
- Report Inappropriate Content
Hello @JanosBecsi ,
There is a part of the macro that is updating numbers with proc SQL update statements using macro variable counts created in a datastep. There are situations where no datasets are actually compared and so none of these numbers are created so the statement would look like:
case
else
Normally with datasets compared there is a when clause for each dataset. When no datasets are compared it can't create any when clauses so it creates the syntax error that you're seeing. I have a newer version that I've made to try to avoid this situation that I will try to upload soon.
@Kurt_Bremser The macro internally uses PROC SQL a lot, which is where the error is coming from after the macro is called.
- Mark as Read
- Mark as New
- Bookmark
- Permalink
- Report Inappropriate Content
Hello @JeffMeyers ,
Did you have time to correct the code? If so, please upload the new version.
Thanks,
Janos
- Mark as Read
- Mark as New
- Bookmark
- Permalink
- Report Inappropriate Content
Hello @JanosBecsi
I have been having trouble uploading new versions of macros to the website. This is where the fixes need to be done in the program if you want to modify yours:
It is basically just adding a when statement that will always be false so it'll never be picked, but if &ndata=0 the case syntax won't resolve in an error.
- Mark as Read
- Mark as New
- Bookmark
- Permalink
- Report Inappropriate Content
Hi Jeff,
if I wanted to compare the the datasets at the subject level what parameters would I update? I am trying to get a report of what information was updated for each subject: what records were added, what records were changed, etc... The macro give a variable summary but is it possible to get this information for each subject record?
Thanks
KC
- Mark as Read
- Mark as New
- Bookmark
- Permalink
- Report Inappropriate Content
Here is the code I wrote before for this purpose.But you need to change it according to your request.
%let old_data=D:\XiaKeShan\卡介苗-MM listing编程需求\LKM-2023-BCG01_study_data_sas_20240117134719\LKM-2023-BCG01_study_data_sas_20240117134719\dataset ; *前一次数据 的路径;
%let new_data=D:\XiaKeShan\卡介苗-MM listing编程需求\LKM-2023-BCG01_study_data_sas_20240129111311\LKM-2023-BCG01_study_data_sas_20240129111311\dataset ; *新数据 的路径;
%let out= D:\XiaKeShan\卡介苗-MM listing编程需求 ; /*指定 输出Excel路径 . 注意:不需要写 want.xlsx */
%let subjid=subject_code; *受试者编号的变量名. 注意:所有的数据集中是一样的,且值是唯一的;
%let drop= SITE_NAME VERSION_NAME PI_NAME USERNAME SUBMIT_TIME UPDATE_USER UPDATE_TIME ; *需要去掉的变量名;
options validvarname=any validmemname=extend mrecall nofmterr nonumber nodate dkricond=nowarn dkrocond=nowarn;
/*******比较 前一次数据 和 新数据********/
libname old v9 "&old_data." access=readonly;
libname new v9 "&new_data." access=readonly;
%macro type_one(idx=,dsn=,title=);
proc datasets library=work kill nolist nodetails;quit;
proc sort data=old.&dsn.(drop=&drop.) out=old;by &subjid. ;run;
proc sort data=new.&dsn.(drop=&drop.) out=new;by &subjid. ;run;
proc compare data=old compare=new noprint out=diff_&dsn.;
id &subjid.;
run;
data call_define;
set diff_&dsn.(drop=_TYPE_ _OBS_);
retain _dummy_ .;
array n{*} _numeric_;
array c{*} _character_;
length vname $ 40;
do i=1 to dim(n);
if n{i} not in (. 0) then do;vname=vname(n{i}); if upcase(vname) not in ("%upcase(&subjid.)") then output;end;
end;
do i=1 to dim(c);
if findc(c{i},'X') then do;vname=vname(c{i}); if upcase(vname) not in ("%upcase(&subjid.)") then output;end;
end;
keep &subjid. vname;
run;
filename t temp;
data _null_;
set call_define;
file t;
/* put 'if &subjid.="' &subjid. '" then call define("' vname '","style","style={background=yellow}");';*/
put "if &subjid.='" &subjid. "' then call define('" vname "','style','style={background=yellow}');";
run;
data &dsn.;
if _n_=1 then do;
length new_change $ 40;
if 0 then set old.&dsn.;
declare hash h(dataset:"old.&dsn.");
h.definekey("&subjid.");
h.definedone();
declare hash h1(dataset:'call_define');
h1.definekey("&subjid.");
h1.definedone();
end;
set new.&dsn.;
new_change='old' ;
if h.check() ne 0 then new_change='new' ;
if h1.check() = 0 then new_change='change' ;
label new_change='数据状态';
run;
/*ods excel file="&out.\&idx..&dsn..xlsx" options(sheet_name="&dsn." autofilter='all' ) ;*/
ods excel options(sheet_name="&title." Embedded_Titles='yes' autofilter='all' ) ;
title j=l link="#目录!A1" "(*ESC*)S={textdecoration=underline color=blue }返回目录";
proc report data=&dsn. split='*' nowd;
column _all_ dummy;
define _all_/display;
define dummy/computed noprint;
compute dummy;
%include t;
endcomp;
run;
/*ods excel close;*/
%mend;
%macro type_two(idx=,dsn=,key=,title=);
proc datasets library=work kill nolist nodetails;quit;
proc sort data=old.&dsn.(drop=&drop.) out=old;by &subjid. &key. ;run;
proc sort data=new.&dsn.(drop=&drop.) out=new;by &subjid. &key. ;run;
proc compare data=old compare=new noprint out=diff_&dsn.;
id &subjid. &key. ;
run;
data call_define;
set diff_&dsn.(drop=_TYPE_ _OBS_);
retain _dummy_ .;
array n{*} _numeric_;
array c{*} _character_;
length vname $ 40;
do i=1 to dim(n);
if n{i} not in (. 0) then do;vname=vname(n{i});if upcase(vname) not in ("%upcase(&subjid.)" "%upcase(&key.)") then output;end;
end;
do i=1 to dim(c);
if findc(c{i},'X') then do;vname=vname(c{i});if upcase(vname) not in ("%upcase(&subjid.)" "%upcase(&key.)") then output;end;
end;
keep &subjid. &key. vname;
run;
filename t temp;
data _null_;
set call_define;
file t;
put "if &subjid.='" &subjid. "' and &key.='" &key. "' then call define('" vname "','style','style={background=yellow}');";
run;
data &dsn.;
if _n_=1 then do;
length new_change $ 40;
if 0 then set new.&dsn.;
if 0 then set old.&dsn.;
declare hash h(dataset:"old.&dsn.");
h.definekey("&subjid.","&key.");
h.definedone();
declare hash h1(dataset:'call_define');
h1.definekey("&subjid.","&key.");
h1.definedone();
end;
set new.&dsn.;
new_change='old' ;
if h.check() ne 0 then new_change='new' ;
if h1.check() = 0 then new_change='change' ;
label new_change='数据状态';
run;
/*ods excel file="&out.\&idx..&dsn..xlsx" options(sheet_name="&dsn." autofilter='all' ) ;*/
ods excel options(sheet_name="&title." Embedded_Titles='yes' autofilter='all' ) ;
title j=l link="#目录!A1" "(*ESC*)S={textdecoration=underline color=blue }返回目录";
proc report data=&dsn. split='*' nowd;
column _all_ dummy;
define _all_/display;
define dummy/computed noprint;
compute dummy;
%include t;
endcomp;
run;
/*ods excel close;*/
%mend;
%macro type_three(idx=,dsn=,key=,key2=,title=);
%local value lower upper ;
%if %upcase(%substr(&dsn.,1,2))=LB %then %do;
proc sql noprint;
select name into :value trimmed from dictionary.columns where libname='OLD' and memname="%upcase(&dsn.)" and label='检查结果';
select name into :lower trimmed from dictionary.columns where libname='OLD' and memname="%upcase(&dsn.)" and label='正常值下限';
select name into :upper trimmed from dictionary.columns where libname='OLD' and memname="%upcase(&dsn.)" and label='正常值上限';
quit;
%put &=value. &=lower. &=upper.;
%end;
proc datasets library=work kill nolist nodetails;quit;
proc sort data=old.&dsn.(drop=&drop.) out=old;by &subjid. &key. &key2.;run;
proc sort data=new.&dsn.(drop=&drop.) out=new;by &subjid. &key. &key2.;run;
proc compare data=old compare=new noprint out=diff_&dsn.;
id &subjid. &key. &key2.;
run;
data call_define;
set diff_&dsn.(drop=_TYPE_ _OBS_);
retain _dummy_ .;
array n{*} _numeric_;
array c{*} _character_;
length vname $ 40;
do i=1 to dim(n);
if n{i} not in (. 0) then do;vname=vname(n{i});if upcase(vname) not in ("%upcase(&subjid.)" "%upcase(&key.)" "%upcase(&key2.)") then output;end;
end;
do i=1 to dim(c);
if findc(c{i},'X') then do;vname=vname(c{i});if upcase(vname) not in ("%upcase(&subjid.)" "%upcase(&key.)" "%upcase(&key2.)") then output;end;
end;
keep &subjid. &key. &key2. vname;
run;
filename t temp;
data _null_;
set call_define;
file t;
put "if &subjid.='" &subjid. "' and &key.='" &key. "' and &key2.='" &key2. "' then call define('" vname "','style','style={background=yellow}');";
run;
data &dsn.;
if _n_=1 then do;
length new_change $ 40;
if 0 then set old.&dsn.;
declare hash h(dataset:"old.&dsn.");
h.definekey("&subjid.","&key.","&key2.");
h.definedone();
declare hash h1(dataset:'call_define');
h1.definekey("&subjid.","&key.","&key2.");
h1.definedone();
end;
set new.&dsn.;
new_change='old' ;
if h.check() ne 0 then new_change='new' ;
if h1.check() = 0 then new_change='change' ;
label new_change='数据状态';
%if %upcase(%substr(&dsn.,1,2))=LB %then %do;
_value=input(&value.,?? best.);
_lower=input(&lower.,?? best.);
_upper=input(&upper.,?? best.);
if not missing(_value) and not missing(_lower) then do; if _value<_lower then &value.=catx(' ',&value.,'-'); end;
if not missing(_value) and not missing(_upper) then do; if _value>_upper then &value.=catx(' ',&value.,'+'); end;
drop _value _lower _upper;
%end;
run;
/*ods excel file="&out.\&idx..&dsn..xlsx" options(sheet_name="&dsn." autofilter='all' ) ;*/
ods excel options(sheet_name="&title." Embedded_Titles='yes' autofilter='all' ) ;
title j=l link="#目录!A1" "(*ESC*)S={textdecoration=underline color=blue }返回目录";
proc report data=&dsn. split='*' nowd;
column _all_ dummy;
define _all_/display;
define dummy/computed noprint;
compute dummy;
%include t;
endcomp;
run;
/*ods excel close;*/
%mend;
/********
注意:
1)目录的值和宏参数(title=)的值保持一致 。
2)目录的值不能包括 /\*~ 等非法字符,因为Sheet名无法显示这些字符。
********/
data 目录;
infile cards truncover expandtabs;
input 序号 目录 $80.;
cards;
1 标题
2 签署知情同意书
3 访视日期
4 人口学资料
5 非膀胱癌既往病史-手术史问询
6 非膀胱癌既往病史-手术史
7 过敏史
8 一般问询
9 膀胱癌病史
10 膀胱癌既往治疗史问询
11 膀胱癌既往治疗史
12 体格检查
13 生命体征问询
14 生命体征
15 ECOG评分
16 胸部X线片
17 超声心动图
18 十二导联心电图
19 C-TST试验
20 血妊娠试验
21 血常规
22 尿常规
23 血生化
24 肌酐
25 凝血功能
26 传染病筛查
27 泌尿系统彩超
28 影像学检查
29 入选排除标准问询
30 入选排除标准
31 筛选结果
32 给药记录表(1周1次)
33 PK采血
34 细胞因子血样采集
35 细胞因子尿样采集
36 药物脱落情况尿样采集
37 不良事件问询
38 不良事件
39 既往或合并用药问询
40 既往或合并用药
41 既往或合并非药物治疗问询
;
ods noresults;
ods _all_ close;
ods excel file="&out.\want.xlsx" ;
ods excel options(sheet_name="目录" Embedded_Titles='yes' ) ;
title "卡介苗-医学监查数据审核表";
proc report data=目录 split='*' nowd;
compute 目录;
call define(_col_,'url',cats('#''',目录,'''!A1') );
call define(_col_,'style','style={textdecoration=underline color=blue}');
endcomp;
run;
%type_one(idx=1,dsn=PRIMARY,title=标题)
%type_one(idx=2,dsn=DS,title=签署知情同意书)
%type_two(idx=3,dsn=SV1,key=folder_name,title=访视日期)
%type_one(idx=4,dsn=DM,title=人口学资料)
%type_one(idx=5,dsn=MH1CO ,title=非膀胱癌既往病史-手术史问询)
%type_two(idx=6,dsn=MH1,key=spid,title=非膀胱癌既往病史-手术史)
%type_one(idx=7,dsn=MH2,title=过敏史)
%type_one(idx=8,dsn=SU,title=一般问询)
%type_one(idx=9,dsn=MH3,title=膀胱癌病史)
%type_one(idx=10,dsn=MH4CO,title=膀胱癌既往治疗史问询)
%type_two(idx=11,dsn=MH4,key=spid,title=膀胱癌既往治疗史)
%type_three(idx=12,dsn=PE,key=folder_name,key2=OID,title=体格检查)
%type_two(idx=13,dsn=VSCO,key=folder_name ,title=生命体征问询)
%type_three(idx=14,dsn=VS,key=spid,key2=OID,title=生命体征)
%type_two(idx=15,dsn=QS,key=folder_name,title=ECOG评分)
%type_two(idx=16,dsn=FA1,key=folder_name,title= 胸部X线片)
%type_two(idx=17,dsn=FA2,key=folder_name,title=超声心动图)
%type_two(idx=18,dsn=EG,key=folder_name,title=十二导联心电图)
%type_two(idx=19,dsn=TST,key=folder_name,title=C-TST试验)
%type_three(idx=20,dsn=LBHCG,key=folder_name,key2=OID ,title=血妊娠试验)
%type_three(idx=21,dsn=LBBR,key=folder_name,key2=OID,title=血常规)
%type_three(idx=22,dsn=LBUR,key=folder_name,key2=OID,title=尿常规)
%type_three(idx=23,dsn=LBBB,key=folder_name,key2=OID,title=血生化)
%type_three(idx=24,dsn=LBSCR,key=folder_name,key2=OID,title=肌酐)
%type_three(idx=25,dsn=LBCG,key=folder_name,key2=OID,title=凝血功能)
%type_two(idx=26,dsn=IFD,key=folder_name,title=传染病筛查)
%type_three(idx=27,dsn=FA3,key=folder_name,key2=OID,title=泌尿系统彩超)
%type_two(idx=28,dsn=FA4,key=folder_name,title=影像学检查)
%type_two(idx=29,dsn=IECO,key=folder_name,title=入选排除标准问询)
%type_two(idx=30,dsn=IE,key=spid,title=入选排除标准)
%type_one(idx=31,dsn=DS1 ,title=筛选结果)
%type_two(idx=32,dsn=EX1,key=OID,title=给药记录表(1周1次))
%type_two(idx=33,dsn=PC,key=OID ,title=PK采血)
%type_two(idx=34,dsn=CB,key=OID,title=细胞因子血样采集)
%type_two(idx=35,dsn=CU,key=OID,title=细胞因子尿样采集)
%type_two(idx=36,dsn=MU,key=OID,title=药物脱落情况尿样采集)
%type_one(idx=37,dsn=AECO,title=不良事件问询)
%type_two(idx=38,dsn=AE,key=spid,title=不良事件)
%type_one(idx=39,dsn=CMCO,title=既往或合并用药问询)
%type_two(idx=40,dsn=CM,key=spid,title=既往或合并用药)
%type_one(idx=41,dsn=CM1CO,title=既往或合并非药物治疗问询)
ods excel close;
- Mark as Read
- Mark as New
- Bookmark
- Permalink
- Report Inappropriate Content
Hello @Kc2
The ID parameter determines what is used to merge and compare rows in datasets. The more ID variables you list the more detailed the comparison gets. The macro is designed to look through the list left to right and only grab the variables that are in the current dataset, so you don't have to worry about finding variables in every dataset.
- Mark as Read
- Mark as New
- Bookmark
- Permalink
- Report Inappropriate Content
Thank you KSharp adn JeffMeyers.
@Ksharp , will test your code.
@JeffMeyers will play with the ID parameters.
April 27 – 30 | Gaylord Texan | Grapevine, Texas
Registration is open
Walk in ready to learn. Walk out ready to deliver. This is the data and AI conference you can't afford to miss.
Register now and save with the early bird rate—just $795!
SAS AI and Machine Learning Courses
The rapid growth of AI technologies is driving an AI skills gap and demand for AI talent. Ready to grow your AI literacy? SAS offers free ways to get started for beginners, business leaders, and analytics professionals of all skill levels. Your future self will thank you.