- Home
- /
- Programming
- /
- ODS Reports
- /
- Re: Selective Horizontal Cell Merging in ExcelXP?
- RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
I'm running SAS 9.4 (9.4 TS1M4) on AIX with Enterprise Guide 7.15 HF2 as a front end. I'm creating a spreadsheet using the ODS ExcelXP tagset. I want to selectively merge cells in columns 6, 7, and 8 together whenever a certain value appears in the cell in column 5.
BOTTOM LINE UP FRONT (with background and code below):
Can SAS with ODS ExcelXP support the generation of XML to selectively merge cells or is it all or nothing? In other words, must I choose between having columns 6, 7, and 8 always merged on every row or can I selectively merge columns 6, 7, and 8 with some rows having the columns merged and some rows with those same columns not merged? XML clearly can support selective merging, but can SAS ODS ExcelXP?
Here's my code. Note the use of the TAGATTR with a "mergeacross".
** If the cell contains a label, bold the cell. **;
** For Payment Pattern 82 (in column 5), merge column 6 across columns 7 and 8. **;
COMPUTE Column6;
IF SUBSTR(Column6,LENGTH(Column6), 1) = ":" THEN
CALL DEFINE ( _col_,'style','style=[FONT=("Calibri",8pt,Bold)]');
ELSE
IF STRIP(Column5) = 'Pmt Pttrn82 25/25/32:' THEN
CALL DEFINE ( _col_,'style','style=[tagattr="mergeacross:2"]');
ENDCOMP;
The following XML is generated if a given row has 'Pmt Pttrn82 25/25/32:' in column 5.
<Row ss:AutoFitHeight="1" ss:Height="14"> <Cell ss:StyleID="data_4" ss:Index="1"><Data ss:Type="String">Pmt Pattern Dt (m):</Data></Cell> <Cell ss:StyleID="data" ss:Index="2"><Data ss:Type="String">08/23/2016 (13)</Data></Cell> <Cell ss:StyleID="data" ss:Index="3"><Data ss:Type="String"></Data></Cell> <Cell ss:StyleID="data" ss:Index="4"><Data ss:Type="String"></Data></Cell> <Cell ss:StyleID="data_5" ss:Index="5"><Data ss:Type="String">Pmt Pttrn82 25/25/32:</Data></Cell> <Cell ss:StyleID="data" ss:MergeAcross="2" ss:Index="6"><Data ss:Type="String">----+----1----+----2----+</Data></Cell> <Cell ss:StyleID="data" ss:Index="7"><Data ss:Type="String"></Data></Cell> <Cell ss:StyleID="data_6" ss:Index="8"><Data ss:Type="String"></Data></Cell> </Row>
Unfortunately, Excel can't open the XML.
If I remove the mergeacross in my SAS program and re-run, Excel can open the SAS generated XML, but of course my cells aren't merged.
When I do the merge manually on the same spreadsheet (after a SAS run with the mergeacross removed), the following XML is created by Excel when I save the spreadsheet:
<Row ss:AutoFitHeight="0" ss:Height="14.0625">
<Cell ss:StyleID="s67"><Data ss:Type="String">Pmt Pattern Dt (m):</Data></Cell>
<Cell ss:StyleID="s68"><Data ss:Type="String">08/23/2016 (13)</Data></Cell>
<Cell ss:StyleID="s68"><Data ss:Type="String"></Data></Cell>
<Cell ss:StyleID="s68"><Data ss:Type="String"></Data></Cell>
<Cell ss:StyleID="s69"><Data ss:Type="String">Pmt Pttrn82 25/25/32:</Data></Cell>
<Cell ss:MergeAcross="2" ss:StyleID="m313941736"><Data ss:Type="String">----+----1----+----2----+</Data></Cell>
</Row>
The XML immediately above saves and re-opens just fine, and the cells are merged the way I want them to be.
Now, you may be saying to yourself "duh, Jim, you have XML for columns 7 and 8 in the SAS generated version." True, but even when I remove the XML for columns 7 and 8, Excel still can't open it. There's something else going on. The XML specifically for the merge (ss:MergeAcross="2") across looks fine. The only difference that I see right off in the row code is that a new StyleID has been introduced in the XML that Excel created after I merged the cells manually.
I also notice that the table tag is different when I save manually vs. what SAS generates.
SAS version:
<Table ss:StyleID="_body">
Excel version:
<Table ss:ExpandedColumnCount="8" ss:ExpandedRowCount="1080" x:FullColumns="1" x:FullRows="1" ss:StyleID="s62" ss:DefaultRowHeight="15">
So, to reiterate the questions posed in the beginning of this post:
Can SAS with ODS ExcelXP support the generation of XML to selectively merge cells or is it all or nothing? In other words, must I choose between having, say, columns 6, 7, and 8 always merged on every row or can I selectively merge columns 6, 7, and 8 with some rows having the columns merged and some rows with those same columns not merged? XML clearly can support selective merging, but can SAS ODS ExcelXP?
Thank you,
Jim
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Have you tried a similar approach with ODS EXCEL instead?
However, I suspect this isn't possible 😞
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Thanks @Reeza,
I suspect you're right that selective horizontal merging isn't possible, at least for now. I can get the mergeacross into the XML, but SAS appears to not be generating whatever accompanying XML is needed to support the mergeacross.
I could try switching from ExcelXP to Excel, but I'm sort of hoping that @Cynthia_sas will weigh in if she has time. She's typically got a pretty good handle on what it is that ODS can and cannot do. I'd hate to burn through a lot of time working on something that just can't be done.
Jim
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Hi:
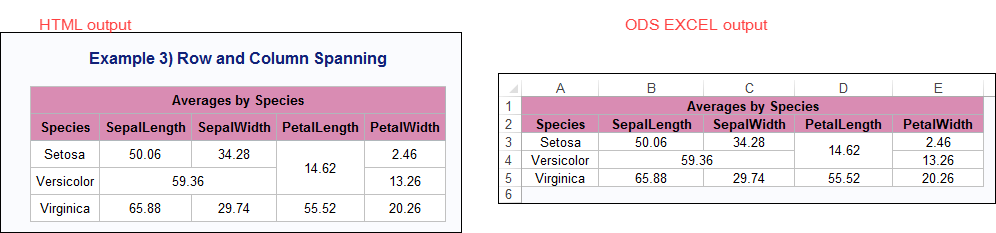
Well, thanks for the vote of confidence. Alas, I do not think you get to there using PROC REPORT. However, while I can't guarantee that ODS EXCEL will use the Report Writing Interface all the time as you want, I was able to modify one of the programs from my "Elvis" paper using SASHELP.IRIS to show both column spanning and row spanning selectively.
Here's the example in HTML compared to ODS EXCEL:
And, here's the code I used:
proc sort data=sashelp.iris out=iris;
by Species;
run;
ods listing close;
ods html path='c:\temp' file="complex_5_iris.html";
ods pdf file="c:\temp\complex_5_iris.pdf";
ods excel file="c:\temp\complex_5_iris.xlsx" ;
title "Example 3) Row and Column Spanning";
data _null_;
set IRIS end=last;
by species;
length longvar $100;
if first.species then do;
spc_cnt = 0;
sl_tot = 0;
sw_tot = 0;
pl_tot = 0;
pw_tot = 0;
end;
spc_cnt + 1;
sl_tot + SepalLength;
sw_tot + sepalwidth;
pl_tot + PetalLength;
pw_tot + PetalWidth;
if last.species then do;
sl_avg = sl_tot / spc_cnt;
sw_avg = sw_tot / spc_cnt;
pl_avg = pl_tot / spc_cnt;
pw_avg = pw_tot / spc_cnt;
end;
if _N_ = 1 then do;
dcl odsout obj();
obj.table_start();
obj.head_start();
** Header row 1;
obj.row_start(type:"Header");
obj.format_cell(text: "Averages by Species", column_span:5, style_attr:"color=black backgroundcolor=CXd98cb3");
obj.row_end();
** Header row 2;
obj.row_start(type: "Header");
obj.format_cell(text: "Species", style_attr:"color=black backgroundcolor=CXd98cb3 fontweight=bold");
obj.format_cell(text: "SepalLength", style_attr:"color=black backgroundcolor=CXd98cb3 fontweight=bold");
obj.format_cell(text: "SepalWidth", style_attr:"color=black backgroundcolor=CXd98cb3 fontweight=bold");
obj.format_cell(text: "PetalLength", style_attr:"color=black backgroundcolor=CXd98cb3 fontweight=bold");
obj.format_cell(text: "PetalWidth", style_attr:"color=black backgroundcolor=CXd98cb3 fontweight=bold");
obj.row_end();
obj.head_end();
end;
** row for every obs;
if last.species then do;
obj.row_start();
if species eq 'Setosa' then do;
obj.format_cell(data: species, row_span:1 );
obj.format_cell(data: sl_avg, row_span:1);
obj.format_cell(data: sw_avg, row_span:1);
obj.format_cell(data: pl_avg, row_span:2,style_attr:"vjust=m");
obj.format_cell(data: pw_avg, row_span:1);
end;
else if species eq 'Versicolor' then do;
obj.format_cell(data: species, row_span:1 );
obj.format_cell(data: sl_avg, row_span:1, column_span:2);
** do not write pl_avg for Versicolor;
obj.format_cell(data: pw_avg, row_span:1);
end;
else if species eq 'Virginica' then do;
obj.format_cell(data: species, row_span:1 );
obj.format_cell(data: sl_avg, row_span:1);
obj.format_cell(data: sw_avg, row_span:1);
obj.format_cell(data: pl_avg, row_span:1);
obj.format_cell(data: pw_avg, row_span:1);
end;
obj.row_end();
end;
if last then do;
obj.table_end();
end;
run;
ods _all_ close; Hope that helps.
Cynthia
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Learn how use the CAT functions in SAS to join values from multiple variables into a single value.
Find more tutorials on the SAS Users YouTube channel.
SAS Training: Just a Click Away
Ready to level-up your skills? Choose your own adventure.