- Home
- /
- Programming
- /
- Graphics
- /
- adjusted model image on proc sgplot
- RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
I have been able to make simple unadusted graphs out of quadtratics in a proc logistic followed by a proc sgplot.
However when i went onto myb adjusted model, we had a disaster:
See image for the result.
What can I do to smooth this out??
ods graphics on;
proc logistic data=HEERDT.VPICU_label plots(only)=effect(x=Ind_SBP1 connect yrange=(0,1) cl );
class Sex_num (ref='0') CABG_i(ref='0') Valve_i(ref='0') Redo1(ref='0') HTN(ref='0') Hyperlip(ref='0') CHF Afib(ref='0')
CM(ref='0') MI(ref='0') AICD(ref='0') DDRF(ref='0') ASA(ref='0') Steroids(ref='0') Amio (ref='0') LMWH(ref='0')
Digoxin(ref='0') AA1(ref='0') Statin1(ref='0')/param =ref;
model VP24_VasoNE_012(ref='0') = Ind_SBP1|Ind_SBP1
Sex_num CABG_i Valve_i Redo1 HTN Hyperlip CHF Afib CM MI AICD DDRF ASA Steroids Amio LMWH Digoxin AA1 Statin1
Age BMI pre_Na pre_BUN pre_Cr pre_Gluc pre_Neutro pre_Lymph pre_Hgb
/ nooddsratio link=glogit ct covb clodds=wald;
output out=QuadSBPVpAdj predicted=Fit lower = l upper = u; /* output predicted values for graphing */
effectplot fit;
where VP24_VasoNE_012 NE 1;
run;
proc sort data =QuadSBPVpAdj ; by Ind_SBP1; run;
proc sgplot data = QuadSBPVpAdj ;
Title "Severe Vasolegia up to 24h by Induction SBP";
band x=Ind_SBP1 lower=l upper=u ;
YAXIS label = "Estimated probably of Severe Vasoplegia in 1st 24 hours";
series x = Ind_SBP1 y = fit/ curvelabel = "Predicted value" lineattrs=(color= Blue pattern=LONGDASHSHORTDASH);;
series x = Ind_SBP1 y = u/ curvelabel = "95% Upper CI" lineattrs= (color=purple pattern=dash);
series x = Ind_SBP1 y = l/ curvelabel = "95% Lower CI" lineattrs = (color =navy pattern = solid thickness =1);
run;
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
You are looking at the predicted probabilities at each of your data points, including the effect of covariates. Use the SCORE statement to get probabilities for a range of Ind_SBP1 values, while keeping covariate values fixed.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
I think I only partially understand this. What would I do with a score or effectplot statement to smooth this curve out?
Even when I add one covariate the curves before very jagged and fuzzy.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
I think you may be plotting the wrong data. Assuming you’re trying to replicate/adjust the EFFECTPLOT you need to use the ODS statement to capture that data as in your previous question.
@ChristosK wrote:
I have been able to make simple unadusted graphs out of quadtratics in a proc logistic followed by a proc sgplot.
However when i went onto myb adjusted model, we had a disaster:
See image for the result.
What can I do to smooth this out??
ods graphics on;
proc logistic data=HEERDT.VPICU_label plots(only)=effect(x=Ind_SBP1 connect yrange=(0,1) cl );class Sex_num (ref='0') CABG_i(ref='0') Valve_i(ref='0') Redo1(ref='0') HTN(ref='0') Hyperlip(ref='0') CHF Afib(ref='0')
CM(ref='0') MI(ref='0') AICD(ref='0') DDRF(ref='0') ASA(ref='0') Steroids(ref='0') Amio (ref='0') LMWH(ref='0')
Digoxin(ref='0') AA1(ref='0') Statin1(ref='0')/param =ref;model VP24_VasoNE_012(ref='0') = Ind_SBP1|Ind_SBP1
Sex_num CABG_i Valve_i Redo1 HTN Hyperlip CHF Afib CM MI AICD DDRF ASA Steroids Amio LMWH Digoxin AA1 Statin1
Age BMI pre_Na pre_BUN pre_Cr pre_Gluc pre_Neutro pre_Lymph pre_Hgb
/ nooddsratio link=glogit ct covb clodds=wald;
output out=QuadSBPVpAdj predicted=Fit lower = l upper = u; /* output predicted values for graphing */
effectplot fit;
where VP24_VasoNE_012 NE 1;
run;
proc sort data =QuadSBPVpAdj ; by Ind_SBP1; run;
proc sgplot data = QuadSBPVpAdj ;
Title "Severe Vasolegia up to 24h by Induction SBP";
band x=Ind_SBP1 lower=l upper=u ;
YAXIS label = "Estimated probably of Severe Vasoplegia in 1st 24 hours";
series x = Ind_SBP1 y = fit/ curvelabel = "Predicted value" lineattrs=(color= Blue pattern=LONGDASHSHORTDASH);;
series x = Ind_SBP1 y = u/ curvelabel = "95% Upper CI" lineattrs= (color=purple pattern=dash);
series x = Ind_SBP1 y = l/ curvelabel = "95% Lower CI" lineattrs = (color =navy pattern = solid thickness =1);
run;
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Reeza:
What do I have to do with an effectplot statement?
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Are you trying to create a customized version of the effect plot? Or something else?
The answers depend on what you're trying to do and we're making assumptions so far...it's easier if you just tell us.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
I was responding to your suggestion and that of PGStats.
Basically I gave a logistic model with a dichotomous outcome. I have a continuous independent variable.
It works better as a spline or a quadratic polynomial rather than as just a linear interpretation(I actually wanted to try out fractional polynomials, but have no idea how to do that).
I have nice curves for the splines and the quadratic, but when I add extra covariates went highly irregular zig zag lines, so thats not presentable. How do I get publication quality adjusted figures? I think I might have got these working before with the sgplot using loess rather than series =x.
Does this make any sense?
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
There’s not an issue with your code but your logic appears flawed. As PG stated, if you don’t set your covariates to constants your prediction’s will vary, after that’s why you included them ,they have a significant effect on the predicted probability.
Fitting lines to predicted data seems weird as well since isn’t that what your model did, predict values? Fitting new lines seems counter counterintuitive unless you’re building a two stage model.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Ok,
So it sounds as though you are saying that with all the adjusting variables in the fulll model, the categorical values should be fixed to one value, and the continuous should be fixed to a mean (or come generally accepted representative value).
I am trying to create two models:
1) just quadratic.
2) Quadratic, and all the confounding variables.
I'm confused here, because if you look at the accompanying output, the first part clearly shows smoothed out predicted values.
I wanted to have more control of the appearance such as having the chance to edit the colours, and to get rid of the gridlines.
Thats the whole point of using proc sgplot. It works fine in just the quadratic model but I don't know what is going on that I need to compensate for in the output from sgplot rather then the logistic step.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
If you look at the plots and the bottom portion specifically, it says what values were selected for the categorical and continuous values and your 'smoothed' plot is the effect plot. The first two plots are almost identical, except one misses some features.
So you can capture the data from that plot using the ODS statements from before and plot that instead of the predicted values at all the different covariate levels.
By default, for effect plots, the mean of continuous values are used and the reference level for the categorical variables are used.
You could choose values that make more sense to you and create your own predictions and plots.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
It looks like all the categorical values in the prior model were having figures drawn as if all their values were 0.
So I tried this instead:
ods graphics on;
%let charvar = Sex_num CABG_i Valve_i Redo1 HTN Hyperlip CHF Afib CM MI AICD DDRF ASA Steroids Amio LMWH Digoxin AA1 Statin1;
proc logistic data =HEERDT.VPICU_label plots(only)=effect(x=Ind_SBP1 connect yrange=(0,1) cl );
class &charvar.;
model VP24_VasoNE_012(ref='0') = Ind_SBP1|Ind_SBP1 Age BMI pre_Na pre_BUN pre_Cr pre_Gluc pre_Neutro pre_Lymph pre_Hgb
&charvar. / nooddsratio ct covb clodds=wald;
output out=QuadSBPVpAdj predicted=Fit lower = l upper = u; /* output predicted values for graphing */
effectplot fit;
where VP24_VasoNE_012 NE 1;
run;
What I was trying to do is see what would happen of I made those categorical values appear to be numeric, and use the macro to set a value for them between 0 and 1.
Looks like the value was taken as 1, and the CI's are so wide now that the output figure is meaningless.
Don't even know what to do if I cant present any adjusted models.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
@ChristosK wrote:
It looks like all the categorical values in the prior model were having figures drawn as if all their values were 0.
Yes, because you specified REF=0 for all of them. There's nothing wrong with that, otherwise why did you set the reference to 0?
@ChristosK wrote:
Looks like the value was taken as 1, and the CI's are so wide now that the output figure is meaningless.
Don't even know what to do if I cant present any adjusted models.
Ok...I don't know what that means, but if your model is meaningless I would assume you'd measure that from other metrics as well, such as the AUC. If your model is not fitting well, that's a problem with the data or how you've built the model. With that many variables I'd expect some inconsistencies. Why not remove insignificant variables and you still need to use param=REF on your CLASS statement for starters? If the model was built with REF=0 I assume that has some clinical significance otherwise pick values that are commonly used. If a variable is continuous don't make it categorical either.
Remember we can't run your code so it's in some ways meaningless to us unless the data is included and even then the odds of someone downloading your data, writing code to import it, running your code, modifying your code and model is really low.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
This is actually quite interesting.
Ive approached this model , and similar outcomes from various different directions.
Have between 15 and 20'ish covariates that I am using, and they are all clinically relevant, and "mostly " still highly significant in the full models.
All of those covariates had a p value well below 0.05 in univariate analysys.
In addition to linear interpretations, I ve run quadratics on the Blood Pressure measures, and splines. And when I made my final tables I compared the odds of specific levels (Centiles 5th, 10th, 25th, 50th, 75th and 95th) compared to the odds at the mode to get my OR's.
In all of these with the fully adjusted spline models the blood pressure measures give significant odds. I have n=1992 patients in my database, and someone else has already published results from a different pool but with n=250k patients.
They published OR curves for splines before and after asjustment, and the adjusted DBP's were still signifiant.
Thats where I am now, Just trying to get meaningful adjusted figures for the quadratic curves or the splines.
Whats interesting to me is that depending on how the previous authors adjusted their models , there may be grounds for questioning their methods . Specifically at what level they adjusted the categorical variables.
Clearly when the categorical values are 0, we get narrowish confidence intervals in then adjusted model.
When the categorical values are 1, the Confidence intervals are extremely wide... so adjusted models are useless perhaps?
Having said that, when I add just one adjusted variable (eg gender or age), we still get the fuzzy lines so for a partially adjusted model we might need a different approach.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
You may create a scoring dataset containing many categorical variables combinations and show the different curves side by side with proc sgpanel.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
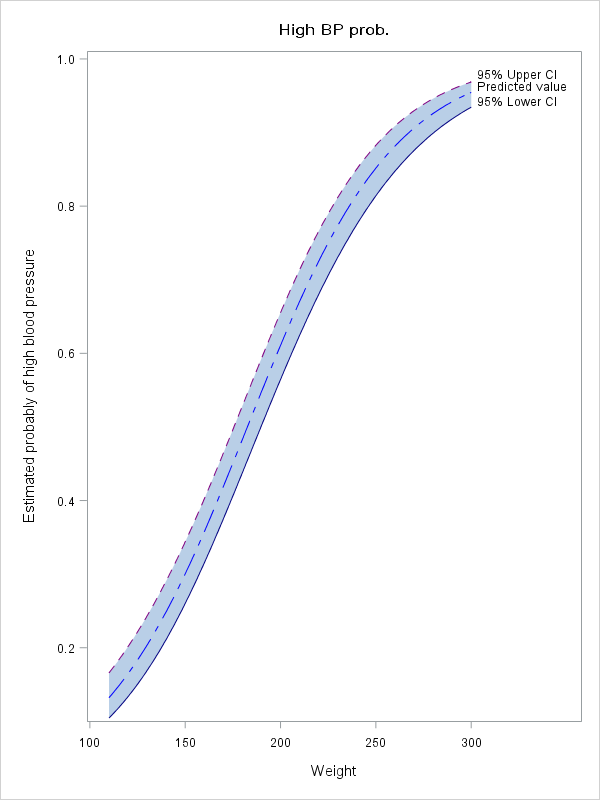
Here is an example of how to do ths with a scoring dataset:
data heart;
set sashelp.heart;
/* Create dichotomous BP status */
if BP_status = "High" then HighBP = "High";
else HighBP = "OK";
run;
data graphScore;
set sashelp.heart(obs=1);
Height = 70;
AgeAtStart = 50;
Cholesterol = 150;
Sex = "Male";
Smoking_Status = "Non-smoker";
do Weight = 110 to 300;
output;
end;
run;
proc logistic data=heart;
class Sex Smoking_Status;
model HighBP = Weight Sex Smoking_Status Height AgeAtStart Cholesterol;
score data=graphScore out=graphPred clm;
run;
ods graphics / height=800 width=600;
proc sgplot data = graphPred ;
Title "High BP prob.";
band x=Weight lower=LCL_High upper=UCL_High ;
YAXIS label = "Estimated probably of high blood pressure";
series x = weight y = P_High/ curvelabel = "Predicted value" lineattrs=(color= Blue pattern=LONGDASHSHORTDASH);;
series x = weight y = UCL_High/ curvelabel = "95% Upper CI" lineattrs= (color=purple pattern=dash);
series x = weight y = LCL_High/ curvelabel = "95% Lower CI" lineattrs = (color =navy pattern = solid thickness =1);
run;
April 27 – 30 | Gaylord Texan | Grapevine, Texas
Registration is open
Walk in ready to learn. Walk out ready to deliver. This is the data and AI conference you can't afford to miss.
Register now and save with the early bird rate—just $795!
Learn how use the CAT functions in SAS to join values from multiple variables into a single value.
Find more tutorials on the SAS Users YouTube channel.
SAS Training: Just a Click Away
Ready to level-up your skills? Choose your own adventure.