- Home
- /
- Analytics
- /
- Stat Procs
- /
- Odds Ratio Calculation for a link different than LOGIT|CLOGIT|GLOGIT
- RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Hi there,
I'm a SAS EG user (7.1 HF1 (7.100.0.2002) (64-bit)).
I have a rather theoretical question concerning the estimation of an odds ratio for a variable involved in an interaction using the GLIMMIX procedure. In this documentation page there is an example on exacly what I would like to do. The example code is:
proc glimmix data=uti;
freq count;
class diagnosis treatment;
model response = diagnosis treatment diagnosis*treatment / dist=binary;
lsmeans diagnosis*treatment / slicediff=diagnosis oddsratio ilink;
run;
In my case, although the response variable is binary, the model is not converging with either the dist=binary (which fixes link=logit by default) or link=logit options. Only when I let the procedure use the default options (dist=gaussian and link=identity) I get it to converge. Maybe the reason for that could be related to the fact that I'm including also a random effect in the model, I don't know.
When I use the example's instructions in my case then, I get the following note:
NOTE: Odds ratios are computed only for the logit, cumulative logit, or the generalized logit link function.
And no odds ratios are outputed.
I have also read this paper and this other one on which they explain how to get the odds ratios through the LSMESTIMATES and EXP option, but they still insist that this is only valid for LINK=LOGIT|CLOGIT|GLOGIT. However, I getting some Exponentiated Estimate column that seems to cointain that I was looking for as it should be the equivalent to the odds ratio.
My question here is, is it correct to assume that the exponentiated estimate corresponds to the odds ratio for models whose link function is different that the previously mentioned LINK=LOGIT|CLOGIT|GLOGIT? Is there any theoretical reason why this is not correct? The fact that such a simple calculation as an exponentiation of a number already computed (the estimate) is not outputed for certain link functions is making me wonder if the reason is because it is a wrong result.
If it is indeed incorrect I need my model to converge with a proper LINK function. Is it a way to make my model converge with LINK=LOGIT? Which are the sensible options for the convergence? What paramenters should I try?
For more details I'm attaching my own code and a piece of the produced output:
proc glimmix data=glimmix_analysis;
class treatarm subjid visitn;
model mainvariable = treatarm visitn treatarm*visitn / solution;
random intercept / subject=subjid solution cl;
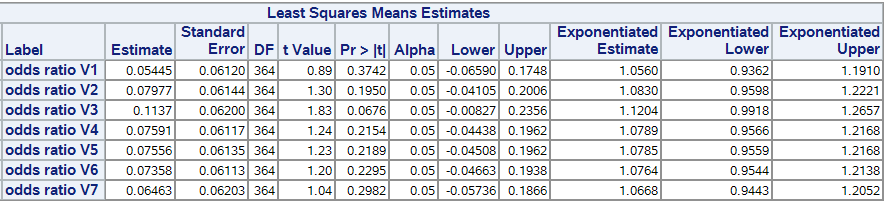
lsmestimate treatarm*visitn "odds ratio V1" 1 0 0 0 0 0 0 -1 0 0 0 0 0 0,
"odds ratio V2" 0 1 0 0 0 0 0 0 -1 0 0 0 0 0,
"odds ratio V3" 0 0 1 0 0 0 0 0 0 -1 0 0 0 0,
"odds ratio V4" 0 0 0 1 0 0 0 0 0 0 -1 0 0 0,
"odds ratio V5" 0 0 0 0 1 0 0 0 0 0 0 -1 0 0,
"odds ratio V6" 0 0 0 0 0 1 0 0 0 0 0 0 -1 0,
"odds ratio V7" 0 0 0 0 0 0 1 0 0 0 0 0 0 -1 / exp cl;
lsmeans treatarm*visitn / slicediff=visitn oddsratio ilink cl;
run;
title;Here is the output of the lsmestimate statement (similar to what I get from the lsmeans except that the exponentiated values are not computed in the other case):
Any hint or clarification will be highly appreciated 🙂
Thanks in advance for your help!!
Accepted Solutions
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
> Is it correct to assume then, if I'm using a different link function (link=identity), that the
> exponentiated estimate can still be interpreted as the Odds Ratio?
No, that is not correct. The odds ratio only make sense when you are comparing the predicted PROBABILITIES for two or more level of classification variables.
When you use DIST=normal and LINK=identity, you are merely fitting a linear model to a response that has values 0 and 1.
The correct way to proceed is to find out why the logistic model is not converging. Please provide any error messages from the log. Also provide the actual code you are using and the output of the NObs table so we can see the size of the sample.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
if seems because a class statement was used, the output format change due to the use of a different estimition mechanism. The documentation for the GLIMMIX procedure clarifies that the exponentiation is the result in this case.
Here is an excerpt from the GLIMMIX Procedure description (red highlight added):
Results designated as odds or odds ratios in the GLIMMIX procedure might reduce to simple exponentiations of solutions in the "Parameter Estimates" table, but they are computed by a different mechanism if the model contains classification variables. The computations rely on general estimable functions; for the MODEL, LSMEANS, and LSMESTIMATEstatements, these functions are based on least squares means. This enables you to obtain odds ratio estimates in more complicated models that involve main effects and interactions, including interactions between continuous and classification variables.
In all cases, the results represent the exponentiation of a linear function of the fixed-effects parameters,
. If
and
are the confidence limits for
on the logit scale, confidence limits for the odds or the odds ratio are obtained as
and
>
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Hi @Cochetti,
Thank you for your quick reponse but I'm afraid I don't fully understand what you mean with your explanation...
Could you comment a little bit more on that?
From what I've read it is more related with the fact of using some specific link functions rather than the kind of variables that you include in your model.
Furthermore, the example that I included in my original post:
http://support.sas.com/kb/24/455.html
has a very similar model to the one that I'm using and the odds ratios are calculated without any problems.
How it is related with having classification variables in your model then?
Thanks again for your patience!
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
I think it's more of a technical thing. GLIMMIX will output results with "Odds Ratio" as the output header when a basic model is generated.
When a more complex model using the class statement is generated, GLIMMIX needs to use a different method to compute the model. The output of this method doesn't label the output in the same way as the basic one, and includes the exponential which can be interpreted as the odds ratio.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Hi @Cochetti,
As I was explaining in my last reply, in this link there is an example of a model with class variables which still writes out the Odds Ratio result without any problem, that's why I thought it is not related to that. Let me paste the example code here:
proc glimmix data=uti;
freq count;
class diagnosis treatment;
model response = diagnosis treatment diagnosis*treatment / dist=binary;
lsmeans diagnosis*treatment / slicediff=diagnosis oddsratio ilink;
run;My case is pretty similar except from the fact that I'm using the default dist=Gaussian (and its corresponding link=identity) because if I use dist=binary (whose default link=logit) my model is not converging.
Let me remind you my original question. In SAS documentation it says (or at least that's what I understand) that you can only calculate the Odds Ratio through the LSMESTIMATE statement and EXP option for the LINK= LOGIT | GLOGIT | CLOGIT. Is it correct to assume then, if I'm using a different link function (link=identity), that the exponentiated estimate can still be interpreted as the Odds Ratio?
Let me quote the SAS documentation page that I'm referring to (its link is here😞
In models with a logit, generalized logit, or cumulative logit link, you can obtain estimates of odds ratios through the ODDSRATIO options in the PROC GLIMMIX, LSMEANS, and MODEL statements. This section provides details about the computation and interpretation of the computed quantities. Note that for these link functions the EXP option in the ESTIMATE and LSMESTIMATE statements also produces odds or odds ratios.
Thanks again for your help and patience.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
> Is it correct to assume then, if I'm using a different link function (link=identity), that the
> exponentiated estimate can still be interpreted as the Odds Ratio?
No, that is not correct. The odds ratio only make sense when you are comparing the predicted PROBABILITIES for two or more level of classification variables.
When you use DIST=normal and LINK=identity, you are merely fitting a linear model to a response that has values 0 and 1.
The correct way to proceed is to find out why the logistic model is not converging. Please provide any error messages from the log. Also provide the actual code you are using and the output of the NObs table so we can see the size of the sample.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Hi @Rick_SAS,
Thank you for such a quick and clear response. That is what I was suspecting.
I wrote my code in my first post, but let me remind it again:
proc glimmix data=glimmix_analysis;
class treatarm subjid visitn;
model mainvariable = treatarm visitn treatarm*visitn / solution;
random intercept / subject=subjid solution cl;
lsmestimate treatarm*visitn "odds ratio V1" 1 0 0 0 0 0 0 -1 0 0 0 0 0 0,
"odds ratio V2" 0 1 0 0 0 0 0 0 -1 0 0 0 0 0,
"odds ratio V3" 0 0 1 0 0 0 0 0 0 -1 0 0 0 0,
"odds ratio V4" 0 0 0 1 0 0 0 0 0 0 -1 0 0 0,
"odds ratio V5" 0 0 0 0 1 0 0 0 0 0 0 -1 0 0,
"odds ratio V6" 0 0 0 0 0 1 0 0 0 0 0 0 -1 0,
"odds ratio V7" 0 0 0 0 0 0 1 0 0 0 0 0 0 -1 / exp cl;
lsmeans treatarm*visitn / slicediff=visitn oddsratio ilink cl;
run;
title;I'm not getting any error/warning in the summary log. I just get a message "Did not converge" in the output. Fyi, I have tried with different optimization techniques with the same result.
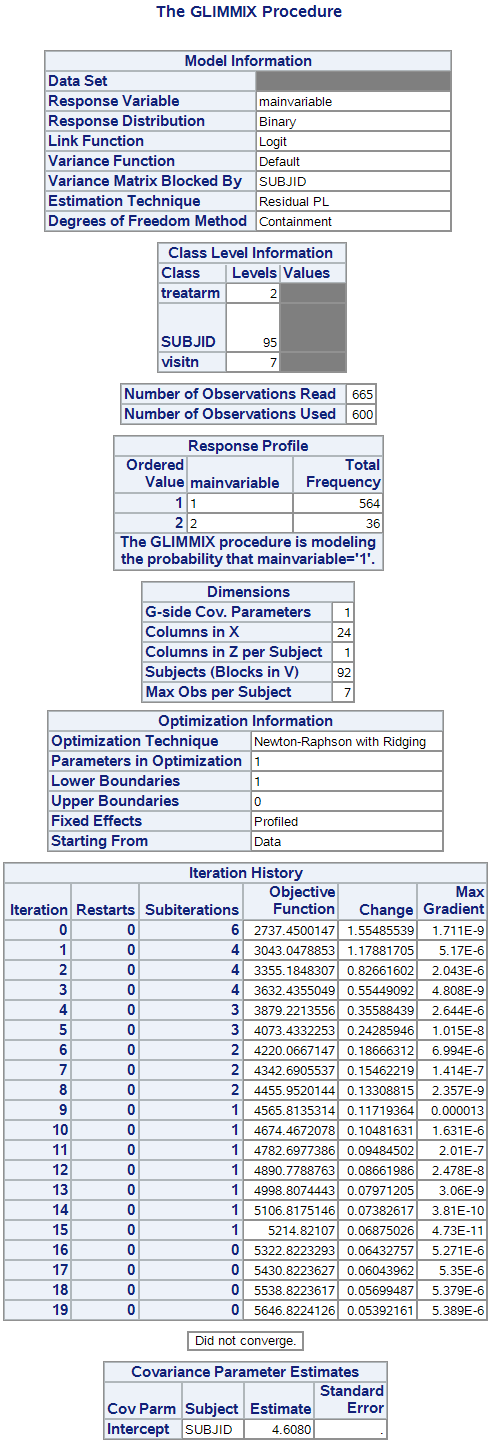
I'm attaching the GLIMMIX output where you can check the sample size and each variable number of levels (I have removed some sensible details). Let me know if there is any other info that I could provide in order to help you detect the problem.
Thank you very much for your help.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Looks like you have 600 nonmissing observations and you are trying to fit a 24-parameter model. Furthermore, you only have 36 obs with mainvariable=2, so there isn't much data to discriminate between event and nonevent. You can see by the "Max Gradient" column in the Iteration History table that the log-likelihood function is very flat, which is why the optimization is not converging to a maximum of the log-likelihood.
Try running just a main effects model (delete the treatarm*visitn effect) and see if you get convergence.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Hi @Rick_SAS,
Yes, I have pretty wierd data. Now it is clear to me why the model is not converging. However, I still have to get some odds ratios from this data. You're right, if I just include only the main effects the model converges, but I need to include somehow the crossing in the model in order to be able to estimate the odds ratios with the lsmestimate or lsmeans statements as you can see in my code. I have tried to include just the interaction term in the model and it is still not converging. Is there any way to get it?
Thanks for your help once again!! 🙂
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
My guess is that there are combinations of treatarm*visitn that do not have any events. Run
proc freq;
where mainvariable=2;
tables treatarm*visitn;
run;
and you will probably see lots of cells that are empty or have only one observation.
If so, the only suggestion I have is to combine some small categories for visitn. For example, if most subjects have 3 visits, you might create a format or a new varialbe that has the values
V = -1 if visitn ❤️
V = 0 if visitn =3
V = 1 if visitn > 3
Or you could make it dichotomous variable:
V = 0 if visitn <=3
V = 1 if visitn > 3
Only by looking at the data can you decide how to combine adjacent categories. But I think then you would be able to examine interactions for the new variable.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Thank you @Rick_SAS,
I cannot regroup the visits the way you are suggesting but I can omit some of them with a lot of missing values from the analysis.
I've seen that my results are very sensitive to these changes so I will have to choose what to do carefully.
However, your tips have been very useful and it's just a matter of testing different options following your guidelines.
Thank you for your help!!
Best regards
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
You could try alternative approaches to the analysis such as a nonmodeling approach with PROC FREQ, a conditional logistic model using the STRATA statement in PROC LOGISTIC, or a Generalized Estimating Equations (GEE) model in PROC GENMOD using the REPEATED statement. As Rick suggests, the problem here is likely that the presence of the interaction makes the data too sparse causing a condition in which some model parameters are infinite and nonexistence of the maximum likelihood solution. PROC LOGISTIC refers to this as "separation" which is described more in this note. The PROC FREQ approach might be particularly attractive for this since you essentially have a stratified 2x2 table with each visit being a stratum. So, as in the note:
proc freq;
table visitn*treatarm*mainvariable / cmh noprint;
run;
Don't miss out on SAS Innovate - Register now for the FREE Livestream!
Can't make it to Vegas? No problem! Watch our general sessions LIVE or on-demand starting April 17th. Hear from SAS execs, best-selling author Adam Grant, Hot Ones host Sean Evans, top tech journalist Kara Swisher, AI expert Cassie Kozyrkov, and the mind-blowing dance crew iLuminate! Plus, get access to over 20 breakout sessions.
ANOVA, or Analysis Of Variance, is used to compare the averages or means of two or more populations to better understand how they differ. Watch this tutorial for more.
Find more tutorials on the SAS Users YouTube channel.