- Home
- /
- Programming
- /
- Graphics
- /
- Re: unending lines in the series plot of proc template
- RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Dear Experts,
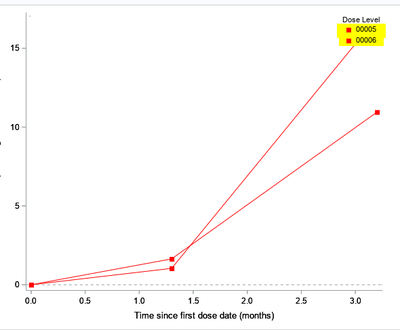
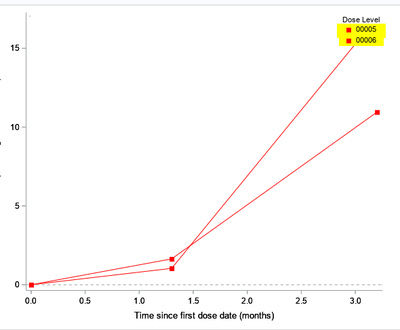
This is with regard to the unending lines issue observed in the proc template when i use the discreteattrmap. I want to use the discreteattrmap and still avoid the unending lines. Here is the sample code and dummy data, please let us know how i can avoid the unending lines.
data full_subset;
input RGROUP$ USUBJID$ AVISITN MONTH CHANGE;
cards;
dose1 00005 1 0 0
dose1 00005 2 1.3 1.639344262
dose1 00005 3 3.2 10.95081967
dose1 00006 1 0 0
dose1 00006 2 1.3 1.039344262
dose1 00006 3 3.2 16.95081967
;
proc sort data=full_subset;
by rgroup usubjid avisitn;
run;
proc template;
define statgraph sgplot;
begingraph / collation=binary dataContrastColors=( red orange green violet yellow grey ) dataSymbols=( EOT );
SymbolChar char=delta_u NAME=EOT / scale=1;
discreteattrmap name='TGROUPS';
value 'Treatment Ongoing' /markerattrs=(symbol=trianglerightfilled color=black size=12);
enddiscreteattrmap;
discreteattrvar attrvar=TGROUP1 var=TGROUP attrmap='TGROUPS';
discreteattrmap name='RGROUPS';
value 'dose1' /markerattrs=(symbol=squarefilled color=red size=12) Lineattrs=( Pattern=1 Thickness=2 color=red);
enddiscreteattrmap;
discreteattrvar attrvar=RGROUP1 var=rgroup attrmap='RGROUPS';
layout overlay / walldisplay=(fill) xaxisopts=( Label="Time since first dose date (months)" type=linear linearopts=(tickvaluesequence=(start=0 end=15 increment=0.5))) y2axisopts=(labelFitPolicy=Split) yaxisopts=( Label="Tumor size (% change from baseline)" labelFitPolicy=Split type=linear ) y2axisopts=(labelFitPolicy=Split);
SeriesPlot X=month Y=change /xaxis=x yaxis=y Group=RGROUP1 LineColorGroup=RGROUP1 MarkerColorGroup=RGROUP1 markersymbolgroup=rgroup1 display=(markers) LegendLabel="CHANGE" NAME="trt";
Layout Gridded / Border=true autoalign=(topleft topright) valign=top;
Layout Gridded;
endlayout;
endlayout;
DiscreteLegend "trt" /*"b"*/ / Location=Inside across=1 type=marker halign=right Title ="Dose Level" ValueAttrs=( Size=5) Opaque=true titleattrs=(family="Arial" size=8pt) titleborder=true sortorder=ascendingformatted autoalign=(TOPRIGHT) order=rowmajor border=false valueattrs=(family="Arial" size=8pt) Opaque=true;
endlayout;
endgraph;
end;
run;
proc sgrender data=FULL_SUBSET template=sgplot ;
run;
What i am expecting is as below, where we can observe the unending lines but the legend is disturbed. I want the legend as dose1 with only 1 row
Please help.
Jag
- Tags:
- graphs
Accepted Solutions
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Hi @Jagadishkatam ,
If you want to just have a different legend, you can simply use a different plot statement without the GROUP option, and then using the attribute option, you can not display the line. Look at the example below, where I use another SERIESPLOT statement to create the legend.
data full_subset;
input RGROUP$ USUBJID$ AVISITN MONTH CHANGE;
cards;
dose1 00005 1 0 0
dose1 00005 2 1.3 1.639344262
dose1 00005 3 3.2 10.95081967
dose1 00006 1 0 0
dose1 00006 2 1.3 1.039344262
dose1 00006 3 3.2 16.95081967
;
proc sort data=full_subset;
by rgroup month;
run;
proc template;
define statgraph sgplot;
begingraph / collation=binary dataContrastColors=( red orange green violet yellow grey ) dataSymbols=( EOT );
SymbolChar char=delta_u NAME=EOT / scale=1;
discreteattrmap name='TGROUPS';
value 'Treatment Ongoing' /markerattrs=(symbol=trianglerightfilled color=black size=12);
enddiscreteattrmap;
discreteattrvar attrvar=TGROUP1 var=TGROUP attrmap='TGROUPS';
discreteattrmap name='RGROUPS';
value 'dose1' /markerattrs=(symbol=squarefilled color=red size=12) Lineattrs=( Pattern=1 Thickness=2 color=red);
enddiscreteattrmap;
discreteattrvar attrvar=RGROUP1 var=rgroup attrmap='RGROUPS';
layout overlay / walldisplay=(fill) xaxisopts=( Label="Time since first dose date (months)" type=linear linearopts=(tickvaluesequence=(start=0 end=15 increment=0.5))) y2axisopts=(labelFitPolicy=Split) yaxisopts=( Label="Tumor size (% change from baseline)" labelFitPolicy=Split type=linear ) y2axisopts=(labelFitPolicy=Split);
SeriesPlot X=month Y=change /xaxis=x yaxis=y Group=USUBJID LineColorGroup=RGROUP1 MarkerColorGroup=RGROUP1 markersymbolgroup=rgroup1 display=(markers);
SeriesPlot X=month Y=change / lineattrs=(thickness=0 color=red) LegendLabel="Dose 1" NAME="trt";
Layout Gridded / Border=true autoalign=(topleft topright) valign=top;
Layout Gridded;
endlayout;
endlayout;
DiscreteLegend "trt" / Location=Inside across=1 type=line halign=right Title ="Dose Level" ValueAttrs=( Size=5) Opaque=true titleattrs=(family="Arial" size=8pt) titleborder=true sortorder=ascendingformatted autoalign=(TOPRIGHT) order=rowmajor border=false valueattrs=(family="Arial" size=8pt) Opaque=true;
endlayout;
endgraph;
end;
run;
proc sgrender data=FULL_SUBSET template=sgplot ;
run;- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
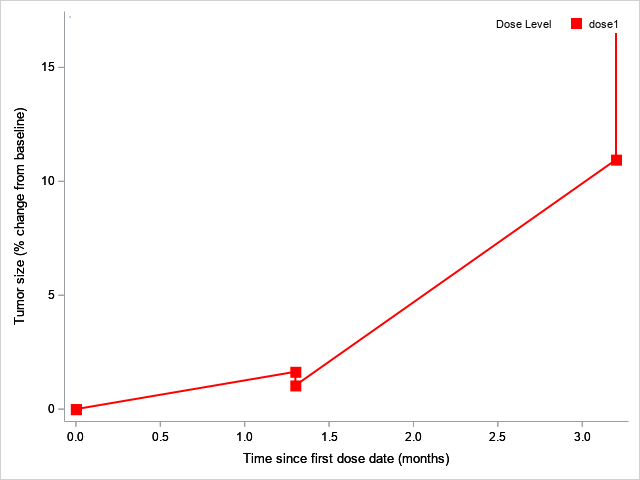
Hi @Jagadishkatam ,
Do you want your image to look like the image below?
If so you use the sort below instead. When using the SERIESPLOT statement it is important to sort the dataset by the value in the x-axis. I noticed you also have two subjects in your dataset, so perhaps you want to use the GROUP=USUBJID option to get two lines, i.e. one for each subject.
proc sort data=full_subset;
by rgroup month;
run;
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Hi @Jagadishkatam,
I've actually just realised that you infact did want to have two separate lines, and so I have put the full code below.
data full_subset;
input RGROUP$ USUBJID$ AVISITN MONTH CHANGE;
cards;
dose1 00005 1 0 0
dose1 00005 2 1.3 1.639344262
dose1 00005 3 3.2 10.95081967
dose1 00006 1 0 0
dose1 00006 2 1.3 1.039344262
dose1 00006 3 3.2 16.95081967
;
proc sort data=full_subset;
by rgroup month;
run;
proc template;
define statgraph sgplot;
begingraph / collation=binary dataContrastColors=( red orange green violet yellow grey ) dataSymbols=( EOT );
SymbolChar char=delta_u NAME=EOT / scale=1;
discreteattrmap name='TGROUPS';
value 'Treatment Ongoing' /markerattrs=(symbol=trianglerightfilled color=black size=12);
enddiscreteattrmap;
discreteattrvar attrvar=TGROUP1 var=TGROUP attrmap='TGROUPS';
discreteattrmap name='RGROUPS';
value 'dose1' /markerattrs=(symbol=squarefilled color=red size=12) Lineattrs=( Pattern=1 Thickness=2 color=red);
enddiscreteattrmap;
discreteattrvar attrvar=RGROUP1 var=rgroup attrmap='RGROUPS';
layout overlay / walldisplay=(fill) xaxisopts=( Label="Time since first dose date (months)" type=linear linearopts=(tickvaluesequence=(start=0 end=15 increment=0.5))) y2axisopts=(labelFitPolicy=Split) yaxisopts=( Label="Tumor size (% change from baseline)" labelFitPolicy=Split type=linear ) y2axisopts=(labelFitPolicy=Split);
SeriesPlot X=month Y=change /xaxis=x yaxis=y Group=USUBJID LineColorGroup=RGROUP1 MarkerColorGroup=RGROUP1 markersymbolgroup=rgroup1 display=(markers) LegendLabel="CHANGE" NAME="trt";
Layout Gridded / Border=true autoalign=(topleft topright) valign=top;
Layout Gridded;
endlayout;
endlayout;
DiscreteLegend "trt" /*"b"*/ / Location=Inside across=1 type=marker halign=right Title ="Dose Level" ValueAttrs=( Size=5) Opaque=true titleattrs=(family="Arial" size=8pt) titleborder=true sortorder=ascendingformatted autoalign=(TOPRIGHT) order=rowmajor border=false valueattrs=(family="Arial" size=8pt) Opaque=true;
endlayout;
endgraph;
end;
run;
proc sgrender data=FULL_SUBSET template=sgplot ;
run;- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Thank you @djrisks for your response.
I tried updating the code to use group=usubjid, and this will generate the graph without the unending lines. But it will affect the legend, like it will display two rows for two subjects and if there are many subjects then the same number of rows will be displayed in the legend.
But this is not my expectation, I want to avoid the unending lines but still display only one row for legend as both the subjects took the same dose i.e., 'dose1'.
Jag
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Hi @Jagadishkatam ,
If you want to just have a different legend, you can simply use a different plot statement without the GROUP option, and then using the attribute option, you can not display the line. Look at the example below, where I use another SERIESPLOT statement to create the legend.
data full_subset;
input RGROUP$ USUBJID$ AVISITN MONTH CHANGE;
cards;
dose1 00005 1 0 0
dose1 00005 2 1.3 1.639344262
dose1 00005 3 3.2 10.95081967
dose1 00006 1 0 0
dose1 00006 2 1.3 1.039344262
dose1 00006 3 3.2 16.95081967
;
proc sort data=full_subset;
by rgroup month;
run;
proc template;
define statgraph sgplot;
begingraph / collation=binary dataContrastColors=( red orange green violet yellow grey ) dataSymbols=( EOT );
SymbolChar char=delta_u NAME=EOT / scale=1;
discreteattrmap name='TGROUPS';
value 'Treatment Ongoing' /markerattrs=(symbol=trianglerightfilled color=black size=12);
enddiscreteattrmap;
discreteattrvar attrvar=TGROUP1 var=TGROUP attrmap='TGROUPS';
discreteattrmap name='RGROUPS';
value 'dose1' /markerattrs=(symbol=squarefilled color=red size=12) Lineattrs=( Pattern=1 Thickness=2 color=red);
enddiscreteattrmap;
discreteattrvar attrvar=RGROUP1 var=rgroup attrmap='RGROUPS';
layout overlay / walldisplay=(fill) xaxisopts=( Label="Time since first dose date (months)" type=linear linearopts=(tickvaluesequence=(start=0 end=15 increment=0.5))) y2axisopts=(labelFitPolicy=Split) yaxisopts=( Label="Tumor size (% change from baseline)" labelFitPolicy=Split type=linear ) y2axisopts=(labelFitPolicy=Split);
SeriesPlot X=month Y=change /xaxis=x yaxis=y Group=USUBJID LineColorGroup=RGROUP1 MarkerColorGroup=RGROUP1 markersymbolgroup=rgroup1 display=(markers);
SeriesPlot X=month Y=change / lineattrs=(thickness=0 color=red) LegendLabel="Dose 1" NAME="trt";
Layout Gridded / Border=true autoalign=(topleft topright) valign=top;
Layout Gridded;
endlayout;
endlayout;
DiscreteLegend "trt" / Location=Inside across=1 type=line halign=right Title ="Dose Level" ValueAttrs=( Size=5) Opaque=true titleattrs=(family="Arial" size=8pt) titleborder=true sortorder=ascendingformatted autoalign=(TOPRIGHT) order=rowmajor border=false valueattrs=(family="Arial" size=8pt) Opaque=true;
endlayout;
endgraph;
end;
run;
proc sgrender data=FULL_SUBSET template=sgplot ;
run;- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Jag
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
I don't know what an "unending line" means.
It looks like the legend is obstructing the lines. If that is the problem, you can
1. Move the legend position
2. Add an offset to the maximum side of the Y axis, such as OFFSETMAX=0.1. See the doc.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
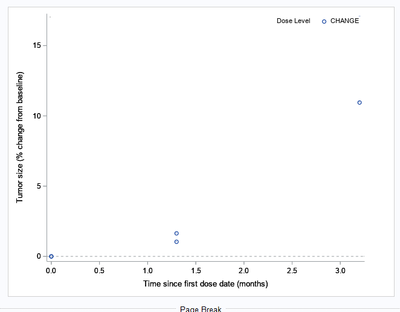
I want to use the group=rgroup1 from discreteattrmap, so that i can use it for controlling the legend.
I hope i am clear, please check the images of the graphs i posted in the main message. Could you please help with me with it.
Jag
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
In your data, after every change in USUBJID, add another observation with missing values for MONTH and CHANGE. Remove the GROUP option from the SERIESPLOT and add BREAK=TRUE to that statement.
Let me know if this works for you.
Thanks!
Dan
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Thank you @DanH_sas for your response ,
I updated the code as below and i get
data full_subset;
infile cards missover;
input
RGROUP$ USUBJID$ AVISITN MONTH CHANGE;
cards;
dose1 00005 1 0 0
dose1 00005 1
dose1 00005 2 1.3 1.639344262
dose1 00005 2
dose1 00005 3 3.2 10.95081967
dose1 00005 3
dose1 00006 1 0 0
dose1 00006 1
dose1 00006 2 1.3 1.039344262
dose1 00006 2
dose1 00006 3 3.2 16.95081967
dose1 00006 3
;
proc sort data=full_subset;
by rgroup usubjid avisitn;
run;
proc template;
define statgraph sgplot;
begingraph / collation=binary dataContrastColors=( red orange green violet yellow grey ) dataSymbols=( EOT );
SymbolChar char=delta_u NAME=EOT / scale=1;
discreteattrmap name='TGROUPS';
value 'Treatment Ongoing' /markerattrs=(symbol=trianglerightfilled color=black size=12);
enddiscreteattrmap;
discreteattrvar attrvar=TGROUP1 var=TGROUP attrmap='TGROUPS';
discreteattrmap name='RGROUPS';
value 'dose1' /markerattrs=(symbol=squarefilled color=red size=12) Lineattrs=( Pattern=1 Thickness=2 color=red);
enddiscreteattrmap;
discreteattrvar attrvar=RGROUP1 var=rgroup attrmap='RGROUPS';
layout overlay / walldisplay=(fill) xaxisopts=( Label="Time since first dose date (months)" type=linear linearopts=(tickvaluesequence=(start=0 end=15 increment=0.5))) y2axisopts=(labelFitPolicy=Split) yaxisopts=( Label="Tumor size (% change from baseline)" labelFitPolicy=Split type=linear ) y2axisopts=(labelFitPolicy=Split);
SeriesPlot X=month Y=change /xaxis=x yaxis=y /*Group=usubjid*/ break=true LineColorGroup=RGROUP1 MarkerColorGroup=RGROUP1 markersymbolgroup=rgroup1 display=(markers) LegendLabel="CHANGE" NAME="trt";
Layout Gridded / Border=true autoalign=(topleft topright) valign=top;
Layout Gridded;
endlayout;
endlayout;
DiscreteLegend "trt" /*"b"*/ / Location=Inside across=1 type=marker halign=right Title ="Dose Level" ValueAttrs=( Size=5) Opaque=true titleattrs=(family="Arial" size=8pt) titleborder=true sortorder=ascendingformatted autoalign=(TOPRIGHT) order=rowmajor border=false valueattrs=(family="Arial" size=8pt) Opaque=true;
endlayout;
endgraph;
end;
run;
proc sgrender data=FULL_SUBSET template=sgplot /*sganno=anno*/;
run;
Jag
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Clearly you need to use GROUP=usubjid to get two separate line segments, one each for "00005" and "00006". Maybe your discreteattrmap and discreteattrvar are not set up correctly. They are set up for the RGROUPS variable, but should be setup for the USUBJID with two values '00005' and '00006'.
discreteattrvar attrvar=USUBJID1 var=usubjid attrmap='USUBJIDS';
Then, use usubjid1 as the group variable.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Jag
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
In that case I suggest you use GROUP=USUBJID, LINECOLORGROUP=RGROUP and MARKERCOLORGROUP=RGROUP. You may be able to use the discreteattrvar RGROUP1 for these two options. You may also need to add the TYPE=LINECOLOR to the Discretelegend.
Also, you may be able to use SGPLOT for this graph with the attrmaps defined as data sets using GROUPLC and LCATTRID.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
For the record, I think you misunderstood my suggestion. You only needed to add one "missing" record after each change in subject ID. This means you should have had one missing record after the last "0005". Having one after the last '0006' would not have hurt, and would make the data step loop simpler.
Thanks!
Dan
SAS Innovate 2025: Save the Date
SAS Innovate 2025 is scheduled for May 6-9 in Orlando, FL. Sign up to be first to learn about the agenda and registration!
Learn how use the CAT functions in SAS to join values from multiple variables into a single value.
Find more tutorials on the SAS Users YouTube channel.
SAS Training: Just a Click Away
Ready to level-up your skills? Choose your own adventure.