- Home
- /
- Analytics
- /
- Stat Procs
- /
- Re: Tukey's HSD Lettered Groupings with Interactions
- RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Hi All,
I am trying to find a way to get the lettered Tukey's groupings for interaction (much like it would on JMP). I've pasted my code below. Currently with the "LSMEANS ... / adjust=tukey pdiff" statement (where the ... would be the different effects I would put in) I get a chart of p-values comparing all the different levels of the interaction. I've look around online and have been told to use "MEANS ... / tukey" statement. When I tried this, the lettered groupings were given for the main effects but not the interactions. Only the mean values of the interactions were given. I found a macro online for doing this called pdmix800, but it only works for Proc Mixed. Any help on this would be greatly appreciated.
proc glm;
class Subject Diameter Height Slope Trial;
model Shld_P1_Abd Shld_P1_Flex Wrist_P1 =
Diameter|Height|Slope subject trial subject*trial
diameter*subject diameter*trial diameter*subject*trial
Height*subject Height*trial Height*subject*trial
Slope*subject Slope*trial Slope*subject*trial/ nouni;
random subject trial subject*trial
diameter*subject diameter*trial diameter*subject*trial
Height*subject Height*trial Height*subject*trial
Slope*subject Slope*trial Slope*subject*trial/ test;
LSMEANS Diameter|Height|Slope / ADJUST=TUKEY PDIFF ;
run;
Accepted Solutions
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
What about the LINES option on the LSMEANS statement.
title1 'Randomized Complete Block With Two Factors';
data PainRelief;
input PainLevel Codeine Acupuncture Relief @@;
datalines;
1 1 1 0.0 1 2 1 0.5 1 1 2 0.6 1 2 2 1.2
2 1 1 0.3 2 2 1 0.6 2 1 2 0.7 2 2 2 1.3
3 1 1 0.4 3 2 1 0.8 3 1 2 0.8 3 2 2 1.6
4 1 1 0.4 4 2 1 0.7 4 1 2 0.9 4 2 2 1.5
5 1 1 0.6 5 2 1 1.0 5 1 2 1.5 5 2 2 1.9
6 1 1 0.9 6 2 1 1.4 6 1 2 1.6 6 2 2 2.3
7 1 1 1.0 7 2 1 1.8 7 1 2 1.7 7 2 2 2.1
8 1 1 1.2 8 2 1 1.7 8 1 2 1.6 8 2 2 2.4
;;;;
run;
proc print;
run;
proc glm data=PainRelief;
class PainLevel Codeine Acupuncture;
model Relief = PainLevel Codeine|Acupuncture;
random painlevel;
lsmeans Codeine|Acupuncture / adjust=tukey lines;
run;
quit;
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
What about the LINES option on the LSMEANS statement.
title1 'Randomized Complete Block With Two Factors';
data PainRelief;
input PainLevel Codeine Acupuncture Relief @@;
datalines;
1 1 1 0.0 1 2 1 0.5 1 1 2 0.6 1 2 2 1.2
2 1 1 0.3 2 2 1 0.6 2 1 2 0.7 2 2 2 1.3
3 1 1 0.4 3 2 1 0.8 3 1 2 0.8 3 2 2 1.6
4 1 1 0.4 4 2 1 0.7 4 1 2 0.9 4 2 2 1.5
5 1 1 0.6 5 2 1 1.0 5 1 2 1.5 5 2 2 1.9
6 1 1 0.9 6 2 1 1.4 6 1 2 1.6 6 2 2 2.3
7 1 1 1.0 7 2 1 1.8 7 1 2 1.7 7 2 2 2.1
8 1 1 1.2 8 2 1 1.7 8 1 2 1.6 8 2 2 2.4
;;;;
run;
proc print;
run;
proc glm data=PainRelief;
class PainLevel Codeine Acupuncture;
model Relief = PainLevel Codeine|Acupuncture;
random painlevel;
lsmeans Codeine|Acupuncture / adjust=tukey lines;
run;
quit;
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Thank you! That was exactly what I needed.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
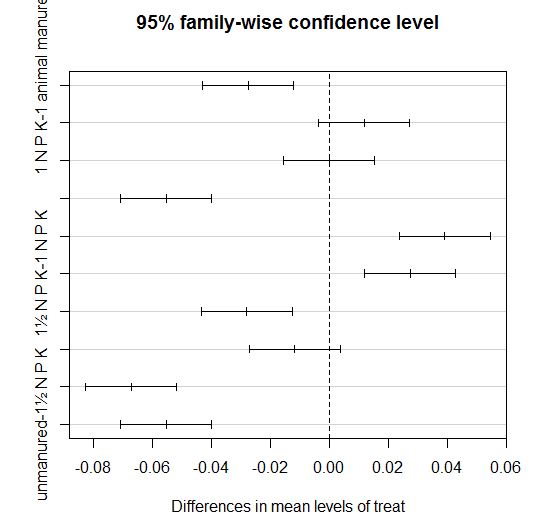
is there any way to obtain graphic tukey separation by means using SAS. I know how to do it in R (PIC BELOW) but I want to learn
how do to it in SAS since most of my code is in SAS.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
I'm not an expert on this, but see the section of the PROC GLM documentation on "Multiple Comparisons." You can choose from about a dozen different multiple comparison adjustments. In particular, read the "Recommendations" and "Interpretation of Multiple Comparisons" sections at the end of the doc.
The main graphical presenation of multiple comparisons is the Hsu diffogram, which is created by default when you have ODS GRAPHICS ON and request multiple comparisons. (Or you can request it with PLOTS=DIFFOGRAM on the MEAN or LSMEANS statement.)
For example, examine the output of
ods graphics on;
proc glm data=sashelp.cars;
class type;
model mpg_city = type;
lsmeans type / adjust=tukey;
run;
For a discussion about how to interpret a diffogram and why many researchers prefer it over the graph that you pasted into your question, see High (2011) "Interpreting the Differences Among LSMEANS in Generalized Linear Models"
Don't miss out on SAS Innovate - Register now for the FREE Livestream!
Can't make it to Vegas? No problem! Watch our general sessions LIVE or on-demand starting April 17th. Hear from SAS execs, best-selling author Adam Grant, Hot Ones host Sean Evans, top tech journalist Kara Swisher, AI expert Cassie Kozyrkov, and the mind-blowing dance crew iLuminate! Plus, get access to over 20 breakout sessions.
ANOVA, or Analysis Of Variance, is used to compare the averages or means of two or more populations to better understand how they differ. Watch this tutorial for more.
Find more tutorials on the SAS Users YouTube channel.