- Home
- /
- Analytics
- /
- Stat Procs
- /
- Proc GLIMMIX for a complex model
- RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Dear all,
I am trying to analyze some data I received after an experiment, and I need your assistance with setting up the correct PROC GLIMMIX syntax.
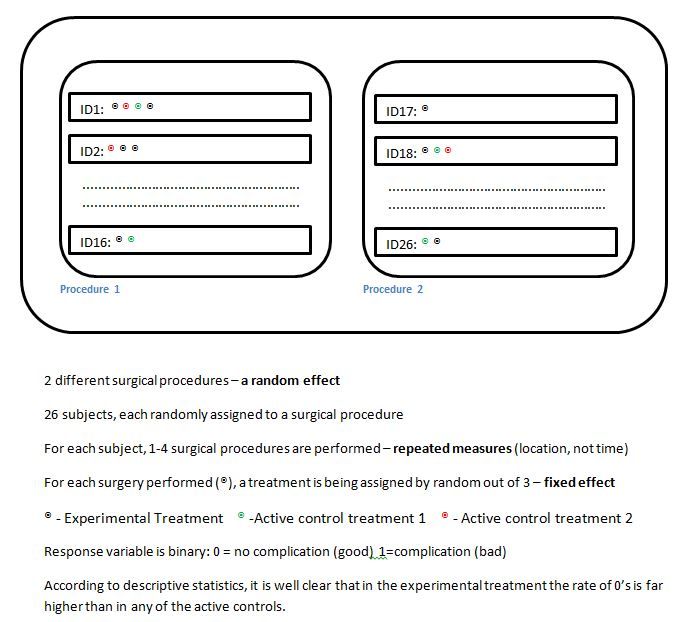
In the attached photo, you can see a scheme I have prepared of what I understand the design is. I will sum the scheme in a few words here as well.
The experiment is dealing with a new experimental surgery tool, that can be used for several surgeries of some kind. The new tool is being tested against 2 current active tools. This is a pilot study, therefore not too much care was taken in designing the experiment, and I think there was no involvement of someone who understands a little bit of statistics.
Two different surgical procedures were used (out of a few possible ones). In total, 26 subjects (animals in this case) were sampled. Each subject was assigned randomly to one and only procedure type. Each subject, went on surgery in 1 or more locations in the body. Each surgery is one unit, or one observation in my data, and each surgery was assigned randomly to one of 3 treatments.
To sum it up, I have:
1 fixed effect - the treatment
1 random effect - the procedure type
1-4 repeated measures per subject
my outcome is binary, 0 means no complication, while 1 means existence of complication. I want to show that the experimental treatment is superior, by having far too 0's than the other two. Originally, the response had levels of complication severity, but due to the small sample, and to the distribution of values, I had no much choice but the merge it and create a binary response (an ordinal model wouldn't work I think).
In the attached photo there is the design as I see it, I have failed to transfer this design into an ANOVA table in the WWFD way (what would Fisher do) way suggest in the book of Walter Stroup, this whole topic of GLMM is new to me and I also struggle with the syntax. I did set something up, but I am very convinced that it's wrong, simply because I still do not understand some basics of the procedure.
My syntax was:
proc glimmix data=data method=quad;
class Subject_ID Treatment Procedure_Type;
model Response = Treatment Treatment(Procedure_Type) / dist = binary link=logit oddsratio;
random intercept / subject=Subject_ID;
random Procedure_Type / subject=Subject_ID;
lsmeans Treatment(Procedure_Type) / slice=Procedure_Type slicediff=Procedure_Type;
run;
I do not understand the structure of the random statement, and have no idea how to tell SAS that I have both a random effect and repeated measures.
This syntax is not more than a "shot in the dark". In the book of Walter Stroup there are more than one way to set up the procedure, I don't understand the very basic difference between (for example):
model Y= a b(a)
model Y= a b a*b
model Y= a|b
Any help with setting up the correct procedure will be most appreciated.
Thank you in advance
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
For random effects like PROCEDURE, use the following version of the RANDOM statement in PROC GLIMMIX:
random PROCEDURE / subject=ID;
For a repeated measure effect like LOCATION, use the following version of the RANDOM statement in PROC GLIMMIX:
random LOCATION / subject=ID residual;
The RESIDUAL option of the RANDOM statement in PROC GLIMMIX converts the effect from a "G-sided" random effect to
an "R-sided" repeated measure.
Fixed effects like TREATMENT should be specified as independent variables in the MODEL statement (to the "right" of the equals sign).
The first MODEL statement you are puzzled about above,
model Y=a b(a);
specifies as independent variables a main effect, a, and a nested effect, b(a), in which the factor, b, is nested within a. Note that this SAS model syntax implies that b does not have a main effect term, independent of its association with a. This MODEL statement also has the equivalent SAS syntax of
model Y=a b*a;
The second and the third MODEL statements you are puzzled about above,
model Y=a b a*b;
and
model Y=a | b;
are equivalent in SAS, with the independent variables in the third statement, a | b, being an abbreviated notation for those in the second statement, a b a*b. Both these MODEL statements specify as independent variables two main effects, a and b, and their interaction, a*b.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Thank you, the differences between the statements are much clearer now.
According to your explanation my MODEL statement is all wrong.
Is there a meaning to defining a random effect AND adding it to the model statement ? From your explanation I conclude that my statement should be:
model Response = Treatment / dist = binary link=logit oddsratio;
random Procedure_Type / subject=Subject_ID;
random location / subject=Subject_ID residual;
I do not have a LOCATION variable. As it happens, I do not know the exact location, all I know is that subject ID1 had 2 surgeries in 2 locations, and let's say ID2 had 3 surgeries in 3 locations, I do not know which locations are they. If I create a new variable representing the repeated measures, we can call it location, than location "1" for ID1, might be a different location than "1" for ID2. What I am saying is that my variables are:
SubjectID, ProcedureType, Treatment, Response
Should I create a new variable representing the repeated measures (what you called LOCATION) ? If so, how will SAS know that I do not mean a time variable with equal spaces ? When defining the residual to move to the "R-Side", do I need to define the type of correlation (AR1, CS, ....) ?
And one more question please, when do I then use:
random intercept / subject=Subject_ID;
Thank you very much, your answer is much clearer than any written guide I found 🙂 .
Edit1: Just tried to run the above statements, and got an error: "R-side random effects are not supported for METHOD=QUAD". I will try LAPLACE and if not will have to settle for PL, which isn't good. What I did was: random _residual_ / subject=Animal_ID type=vc;
which is similar to adding residual at the end of the statement with intercept at the beginning (I am learning slowly 🙂 )
Edit2: I ran the lsmeans statement: lsmeans Treatment / slice=Procedure_Type slicediff=Procedure_Type plot=meanplot(sliceby=Procedure_Type);
not only that it ain't slicing by Procedure_Type like I want, it gives me negative estimators for the 3 treatments. Maybe I miss something basic, but how can values of 0 and 1 yield a negative mean ?
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
My suggestions about your earlier questions are in the attached .TXT document.
The LSMEANS estimates for treatment are negative because of your use of the logit link, where probabilities between 0 and 1 are specified as logits, ln(p/(1-p)), which can be less than zero when the estimated probability is less than 0.50 [for example, if p=0.40, ln(0.4/0.6) ~= - 0.41].
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
One other point--with a binomial distribution, using the residual option means that you are fitting a marginal model, where the estimated proportions are "regressed toward the mean", as opposed to a conditional model where the estimated proportions are those expected values given the random effects. So on this I disagree some with Matt, and I would fit:
proc glimmix data=data method=quad;
class Subject_ID Treatment Procedure_Type;
model Response = Treatment|Procedure_Type / dist = binary link=logit oddsratio;
random intercept / subject=Subject_ID;
random Treatment/subject=subject_id type= <pick between CSH, CHOL, or VC>;
lsmeans Treatment*Procedure_Type / slice=Procedure_Type slicediff=Procedure_Type ilink oddsratio;
run;
Here I fit the repeated measures on the G-side, and choose between three covariance structures having unequal values on the diagonal, and either zeros for covariance (VC), a constant covariance factor (CSH), or a fully unstructured covariance (CHOL).
Steve Denham
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Thank you both for your help.
Matt, thank you for the long explanation, it is very informative !
Steve, in your code the Procedure_Type is a fixed effect, did you mean in the 3rd line of the code to write Procedure_Type, or should I add another random statement ?
When I ran the code, the G matrix wasn't positive defined, could it be because I had 2 random statements with Subject_ID ?
I also got weird OR estimated:
| OR | DF | L_CL | U_CL | ||||
| Treatment1 | Treatment3 | <0.001 | 11 | <0.001 | <0.001 | ||
|---|---|---|---|---|---|---|---|
| Treatment2 | Treatment3 | <0.001 | 11 | <0.001 | <0.001 | ||
| Procedure1 | Procedure2 | >999.999 | 26 | 440.053 | >999.999 |
| Effect | NUM DF | DEN DF | F | P.V |
| Treatment | 2 | 11 | 39.32 | <.0001 |
|---|---|---|---|---|
| Procedure_Type | 1 | 26 | 45.09 | <.0001 |
| Treatment*Procedure_ | 2 | 26 | Infty | <.0001 |
When I ran Matt's code I have also received some wide confidence intervals for OR, however the F value did not converged to infinity (I did not have Procedure_Type as a fixed effect)
Can these outputs be a result of what Matt has mentioned, the model balking due to the fact that some subjects had 2 surgeries with the same treatment, which are my repeated measures ?
Some more outputs:
Pearson Chi-Square / DF = 0.69 (which is good)
| covariance prarameter estimates | |||
| Intercept | Subject_ID | 0.5162 | 1.4348 |
|---|---|---|---|
| Treatment | Subject_ID | 1.14E-13 | . |
Edit1: When I changed Steve's code and replaced the Subject_ID by Procedure_Type in the random statement, I have received the following error message:
Estimation by quadrature is available only if the data can be processed by subjects.
Make sure that all G-side RANDOM statements have SUBJECT= effects. If there are multiple
SUBJECT= effects they need to form a containment hierarchy, e.g., SUBJECT=A,
SUBJECT=A*B, SUBJECT=A(B), ... .
Edit 2: When I removed the interaction term (which wasn't significant) I got slightly more reasonable results, the code was:
proc glimmix data=data method=quad;
class Subject_ID Treatment Procedure_Type;
model Response = Treatment Procedure_Type / dist = binary link=logit oddsratio;
random Treatment / subject=Subject type=vc;
run;
By the way, silly question maybe, how do I interpret an OR value of less than 1 ? I always find it hard (treatment 3 has many more 0's - non events, than treatments 1 and 2)
For example if I get
Treatment Treatment OR
1 3 0.009
Edit 3: I have "re-coded" the response, to get OR larger than 1, and I get an OR of 60 with CI of 6 to 600, due to small sample and perhaps other factors, how can I report such results ? I mean, it is significant (1 is not inside the CI), but I am not being very precise, am I ? One of the reasons for this is the sample size, in the control treatments, there are hardly any "successes". I have 15 samples in one of them, with 14 failures and 1 success, so the "1" is the problem.
Don't miss out on SAS Innovate - Register now for the FREE Livestream!
Can't make it to Vegas? No problem! Watch our general sessions LIVE or on-demand starting April 17th. Hear from SAS execs, best-selling author Adam Grant, Hot Ones host Sean Evans, top tech journalist Kara Swisher, AI expert Cassie Kozyrkov, and the mind-blowing dance crew iLuminate! Plus, get access to over 20 breakout sessions.
ANOVA, or Analysis Of Variance, is used to compare the averages or means of two or more populations to better understand how they differ. Watch this tutorial for more.
Find more tutorials on the SAS Users YouTube channel.



