- Home
- /
- Analytics
- /
- Stat Procs
- /
- Re: PROC GLIMMIX: picking correct model using "Fit Statistics"
- RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Accepted Solutions
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
For the normal and lognormal distributions, the IC values can be used to compare different models, at least as far as the covariance structure selection is concerned. For models differing in the fixed effects, a likelihood ratio test is my preferred tool.
For these two models, what differences in the models and parameterization are you looking at?
Steve Denham
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
For the normal and lognormal distributions, the IC values can be used to compare different models, at least as far as the covariance structure selection is concerned. For models differing in the fixed effects, a likelihood ratio test is my preferred tool.
For these two models, what differences in the models and parameterization are you looking at?
Steve Denham
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Feedlot Trial with 4 trt (experimental unit = animal). Collected blood 3 times (diff. days) and analyzed 21 blood serum components each time. All blood data are continuous, even though some variables like ALT (raw data below) look like count data (due to what the machine spits out).
EDITOR (from your previous help):
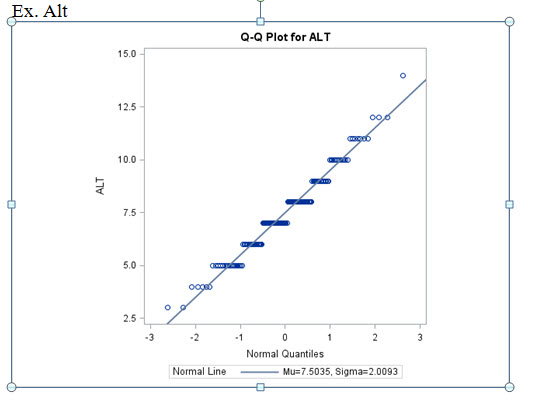
using "dist=lognormal" for non-normal data (looking at Q-Q plots, Shap./Wilk) and dist=normal for norm data. I'm also trying to fit the best structure (like in PROC MIXED), using CS, CSH, AR(1), and ARH(1).
PROC GLIMMIX;
CLASS ID TRT DAY;
MODEL ALT = TRT day trt*day/dist=normal ddfm=kr solution;
Random day /residual subject = ID(trt) type =CSH;
LSMEANS TRT day/DIFF ADJUST=simulate;
LSmeans trt*day/slicediff=day adjust=SIMULATE adjdfe=row
QUESTIONS (if answered, will save me a lot of time / headaches😞
- Is PROC MIXED and PROC GLIMMIX with dist=normal, the same thing (if variable is normal)? I know about the repeated statement in MIXED vs. random residual statement in GLIMMIX.
- Is transforming data (lognormal) & running PROC Mixed the same thing as PROC GLIMMIX with dist = lognormal?
- Since all my serum data are continuous, I think the only for distribution options I have available are lognormal, beta and gamma, correct? Or, can discrete distribution options (neg binomial, poisson, etc.) be used for continuous data?
*if so, then how to you determine best fit (AIC, BIC, still OK to use, when comparing these distribution options with Lognormal & normal?
| ID | TRT | PHASE | DAY | ALT | GGT | |
| 1 | RED | 1 | 0 | 6 | 52 | |
| 1 | RED | 1 | 14 | 10 | 42.1 | |
| 1 | RED | 2 | 57 | 9 | 40.8 | |
| 2 | CSH | 1 | 0 | 6 | 58.5 | |
| 2 | CSH | 1 | 14 | 7 | 60.1 | |
| 2 | CSH | 2 | 57 | 7 | 61 | |
| 3 | BLU | 1 | 0 | 6 | 58.3 | |
| 3 | BLU | 1 | 14 | 9 | 52.6 | |
| 3 | BLU | 2 | 57 | 7 | 57 | |
| 4 | BLU | 1 | 0 | 10 | 43.3 | |
| 4 | BLU | 1 | 14 | 11 | 42.2 | |
| 4 | BLU | 2 | 57 | 7 | 38.3 | |
| 6 | ONE | 1 | 0 | 4 | 58.6 | |
| 6 | ONE | 1 | 14 | 7 | 60.1 | |
| 6 | ONE | 2 | 57 | 8 | 56.5 | |
| 7 | BLU | 1 | 0 | 7 | 41.5 | |
| 7 | BLU | 1 | 14 | 8 | 36.3 | |
| 7 | BLU | 2 | 57 | 6 | 41.5 | |
| 8 | BLU | 1 | 0 | 5 | 55.8 | |
| 8 | BLU | 1 | 14 | 8 | 51.9 | |
| 8 | BLU | 2 | 57 | 6 | 44.1 | |
| 10 | BLU | 1 | 0 | 6 | 41.7 | |
| 10 | BLU | 1 | 14 | 11 | 42.4 | |
| 10 | BLU | 2 | 57 | 10 | 43.5 | |
| 11 | RED | 1 | 0 | 8 | 52.4 | |
| 11 | RED | 1 | 14 | 9 | 50.1 | |
| 11 | RED | 2 | 57 | 7 | 51.7 | |
| 12 | ONE | 1 | 0 | 8 | 46.6 | |
| 12 | ONE | 1 | 14 | 11 | 49 | |
| 12 | ONE | 2 | 57 | 8 | 53.3 | |
| 13 | ONE | 1 | 0 | 5 | 56.1 | |
| 13 | ONE | 2 | 57 | 7 | 53.3 | |
| 14 | CSH | 1 | 0 | 5 | 60.1 | |
| 14 | CSH | 1 | 14 | 5 | 55.3 | |
| 14 | CSH | 2 | 57 | 4 | 56 | |
| 15 | CSH | 1 | 0 | 8 | 47.9 | |
| 15 | CSH | 1 | 14 | 9 | 55.8 | |
| 15 | CSH | 2 | 57 | 4 | 53.1 | |
| 16 | ONE | 1 | 0 | 6 | 66.1 | |
| 16 | ONE | 1 | 14 | 7 | 54.8 | |
| 16 | ONE | 2 | 57 | 8 | 58 | |
| 18 | RED | 1 | 0 | 8 | 41.8 | |
| 18 | RED | 1 | 14 | 10 | 40.9 | |
| 18 | RED | 2 | 57 | 7 | 42.8 | |
| 19 | ERC | 1 | 0 | 3 | 53 | |
| 19 | ERC | 1 | 14 | 5 | 46.7 | |
| 19 | ERC | 2 | 57 | 5 | 43.4 | |
| 20 | RED | 1 | 0 | 9 | 48.8 | |
| 20 | RED | 1 | 14 | 9 | 41.3 | |
| 20 | RED | 2 | 57 | 6 | 48.6 | |
| 22 | RED | 1 | 0 | 7 | 46.2 | |
| 22 | RED | 1 | 14 | 10 | 44.2 | |
| 22 | RED | 2 | 57 | 8 | 44 | |
| 24 | ERC | 1 | 0 | 5 | 36.5 | |
| 24 | ERC | 1 | 14 | 8 | 41.7 | |
| 24 | ERC | 2 | 57 | 14 | 36.9 | |
| 25 | ERC | 1 | 0 | 8 | 56.4 | |
| 25 | ERC | 1 | 14 | 9 | 57.7 | |
| 25 | ERC | 2 | 57 | 8 | 48.4 | |
| 67 | ONE | 1 | 0 | 7 | 64.8 | |
| 67 | ONE | 1 | 14 | 9 | 63.1 | |
| 67 | ONE | 2 | 57 | 5 | 53.3 | |
| 71 | MESQ | 1 | 0 | 5 | 59.8 | |
| 71 | MESQ | 1 | 14 | 8 | 51.9 | |
| 71 | MESQ | 2 | 57 | 7 | 57 | |
| 72 | CSH | 1 | 0 | 7 | 53.9 | |
| 72 | CSH | 1 | 14 | 6 | 56.4 | |
| 72 | CSH | 2 | 57 | 6 | 60.1 | |
| 75 | RED | 1 | 0 | 6 | 49.8 | |
| 75 | RED | 1 | 14 | 9 | 49.4 | |
| 75 | RED | 2 | 57 | 6 | 43.3 | |
| 76 | MESQ | 1 | 0 | 6 | 48.6 | |
| 76 | MESQ | 1 | 14 | 8 | 44.7 | |
| 76 | MESQ | 2 | 57 | 7 | 46.4 | |
| 78 | ERC | 1 | 0 | 9 | 65.4 | |
| 78 | ERC | 1 | 14 | 9 | 67.6 | |
| 79 | MESQ | 1 | 0 | 8 | 39.6 | |
| 79 | MESQ | 1 | 14 | 12 | 44.4 | |
| 79 | MESQ | 2 | 57 | 6 | 39.2 | |
| 80 | ONE | 1 | 0 | 8 | 46.2 | |
| 80 | ONE | 1 | 14 | 8 | 51.1 | |
| 80 | ONE | 2 | 57 | 7 | 46.8 | |
| 81 | ERC | 1 | 0 | 9 | 52.1 | |
| 81 | ERC | 1 | 14 | 10 | 54.6 | |
| 81 | ERC | 2 | 57 | 10 | 52.9 | |
| 83 | RED | 1 | 0 | 9 | 44.3 | |
| 83 | RED | 1 | 14 | 8 | 42 | |
| 84 | RED | 1 | 0 | 7 | 68.5 | |
| 84 | RED | 1 | 14 | 10 | 60.5 | |
| 84 | RED | 2 | 57 | 12 | 50.9 | |
| 85 | CSH | 1 | 0 | 8 | 57.2 | |
| 85 | CSH | 1 | 14 | 9 | 59.2 | |
| 85 | CSH | 2 | 57 | 7 | 51.7 | |
| 86 | MESQ | 1 | 0 | 8 | 40.3 | |
| 86 | MESQ | 1 | 14 | 10 | 40.2 | |
| 86 | MESQ | 2 | 57 | 7 | 46.4 | |
| 87 | CSH | 1 | 0 | 5 | 50 | |
| 87 | CSH | 1 | 14 | 6 | 51.4 | |
| 87 | CSH | 2 | 57 | 5 | 44.7 | |
| 88 | CSH | 1 | 0 | 7 | 48.2 | |
| 88 | CSH | 1 | 14 | 7 | 54.4 | |
| 88 | CSH | 2 | 57 | 5 | 51.9 | |
| 89 | MESQ | 1 | 0 | 5 | 43.5 | |
| 89 | MESQ | 1 | 14 | 8 | 45.8 | |
| 89 | MESQ | 2 | 57 | 5 | 43.1 | |
| 90 | BLU | 1 | 0 | 9 | 49.2 | |
| 90 | BLU | 1 | 14 | 8 | 54.7 | |
| 90 | BLU | 2 | 57 | 6 | 47.4 | |
| 91 | ONE | 1 | 0 | 8 | 50.9 | |
| 91 | ONE | 1 | 14 | 7 | 49.2 | |
| 91 | ONE | 2 | 57 | 5 | 49.9 | |
| 93 | BLU | 1 | 0 | 7 | 35.8 | |
| 93 | BLU | 1 | 14 | 11 | 33.5 | |
| 93 | BLU | 2 | 57 | 10 | 39.2 | |
| 94 | MESQ | 1 | 0 | 7 | 50.2 | |
| 94 | MESQ | 1 | 14 | 8 | 57.5 | |
| 94 | MESQ | 2 | 57 | 7 | 59.5 | |
| 95 | ONE | 1 | 0 | 8 | 49.9 | |
| 95 | ONE | 1 | 14 | 11 | 46.8 | |
| 95 | ONE | 2 | 57 | 8 | 44.6 | |
| 96 | ERC | 1 | 0 | 6 | 62.1 | |
| 96 | ERC | 1 | 14 | 12 | 59.6 | |
| 96 | ERC | 2 | 57 | 7 | 61.1 | |
| 97 | MESQ | 1 | 0 | 4 | 50.1 | |
| 97 | MESQ | 1 | 14 | 7 | 46.2 | |
| 97 | MESQ | 2 | 57 | 3 | 48.7 | |
| 98 | BLU | 1 | 0 | 7 | 68.8 | |
| 98 | BLU | 1 | 14 | 11 | 65.6 | |
| 98 | BLU | 2 | 57 | 7 | 62.2 | |
| 99 | ERC | 1 | 0 | 8 | 53.1 | |
| 99 | ERC | 1 | 14 | 11 | 49.1 | |
| 99 | ERC | 2 | 57 | 10 | 45.1 | |
| 100 | ERC | 1 | 0 | 7 | 53.6 | |
| 100 | ERC | 1 | 14 | 9 | 57.6 | |
| 100 | ERC | 2 | 57 | 10 | 54 | |
| 4052 | MESQ | 1 | 0 | 4 | 46.1 | |
| 4052 | MESQ | 1 | 14 | 8 | 44.6 | |
| 4052 | MESQ | 2 | 57 | 6 | 53 | |
| 4053 | CSH | 1 | 0 | 5 | 51.6 | |
| 4053 | CSH | 1 | 14 | 6 | 66.3 | |
| 4053 | CSH | 2 | 57 | 5 | 53.8 |
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Sorry to be slow getting back to this forum, but have had meetings to attend.
- Is PROC MIXED and PROC GLIMMIX with dist=normal, the same thing (if variable is normal)? I know about the repeated statement in MIXED vs. random residual statement in GLIMMIX.
Yes, although the optimization method may differ, so that you get slightly different results.
- Is transforming data (lognormal) & running PROC Mixed the same thing as PROC GLIMMIX with dist = lognormal?
Yes--think about what a lognormal distribution is: that the log(estimated value) have an error that is normal(mu, sigma**2). This is what you expect with log transformed data. Just remember that exponentiating the returned lsmeans/estimates does NOT give you the expected value on the original scale, but rather the expected median.
- Since all my serum data are continuous, I think the only for distribution options I have available are lognormal, beta and gamma, correct? Or, can discrete distribution options (neg binomial, poisson, etc.) be used for continuous data?
Well, you can use them, just beware of what is going on with assumptions regarding expected values and variances, etc. For instance, a Poisson assumes that the means and variances are equal, and the optimization tries to take that into account. Also, recall that the beta distribution is only defined on the interval [0,1], so unless you scale all of your data to that range, it really is probably not a candidate. Other continuous distributions with large skew include exponential (special case of the gamma) and inverse Gaussian (or Wald).
*if so, then how to you determine best fit (AIC, BIC, still OK to use, when comparing these distribution options with Lognormal & normal?
This is harder, because you can't compare the IC for one distribution to another--they are dependent on the log likelihood of the data, which is going to be in turn dependent on the link function used in GLIMMIX. Choosing the "best" distribution should be done based more on the processes involved in the generation of the data than on anything else. In this case you have serum data. You know it is bounded below by zero, and has a long tail to the right. That defines a lognormal distribution (and a Wald and a gamma). Selection should be based on things like the CV--is it constant for different levels of the independent variable? If so, probably lognormal. If not, maybe gamma. If you are really determined to look at the "best" fit, try plotting the observed values vs. predicted values, and look for any systematic deviations from a straight line.
Steve Denham
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
As always, that helps alot. Just a little confused on your statement "Just remember that exponentiating the returned lsmeans/estimates does NOT give you the expected value on the original scale, but rather the expected median..."
When I run GLIMMIX dist = lognormal, then backtransform (code below), those backtransformed values (LSmeans and SEM) are what I'd report in a publication, correct?
PROC GLIMMIX;
CLASS ID TRT DAY;
MODEL CPK = TRT day trt*day/dist=lognormal ddfm=kr solution;
Random day /residual subject = ID(trt) type =CSH;
LSMEANS TRT day/DIFF ADJUST=simulate;
LSmeans trt*day/slicediff=day adjust=SIMULATE adjdfe=row;
ODS OUTPUT lsmeans=lsmeans;
PROC PRINT; RUN; quit;
data btlsmeans;
set lsmeans;
omega=exp(stderr*stderr);
btlsmean=exp(estimate)*sqrt(omega);
btvar=exp(2*estimate)*omega*(omega-1);
btsem=sqrt(btvar);
PROC PRINT; RUN;
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
This
data btlsmeans;
set lsmeans;
omega=exp(stderr*stderr);
btlsmean=exp(estimate)*sqrt(omega);
btvar=exp(2*estimate)*omega*(omega-1);
btsem=sqrt(btvar);
PROC PRINT; RUN;gives you what should be reported for lognormal data.
This:
data btlsmeans;
set lsmeans;
btlsmean=exp(estimate);
PROC PRINT; RUN;gives the median. And exponentiating the standard error gives something uninterpretable.
So, you are doing everything you need for publication type tables.
Steve Denham
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Steve.
I have some percentage data that needs to be transformed:
ARSIN(SQRT(X))
2 questions:
1. Do I need to divide "X" above, by anything? I saw an article that suggested you divide by number per group, but it wasn't very clear.
2. Do you have a "handy" backtransformed code for ARSIN Like you did for log?
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
As I get caught up from time spent hobnobbing with my fellow wizards (ref. L. Frank Baum, The Wizard of OZ), I think I keep throwing this out there. Don't use the arcsine square root transform. Use the proper distribution in glimmix. If the variable is a ratio of two continuous random variables, then a beta distribution makes the most sense. If the variable is a true proportion (summed up binary yes/no), then binomial is the best bet.
Steve Denham
Don't miss out on SAS Innovate - Register now for the FREE Livestream!
Can't make it to Vegas? No problem! Watch our general sessions LIVE or on-demand starting April 17th. Hear from SAS execs, best-selling author Adam Grant, Hot Ones host Sean Evans, top tech journalist Kara Swisher, AI expert Cassie Kozyrkov, and the mind-blowing dance crew iLuminate! Plus, get access to over 20 breakout sessions.
ANOVA, or Analysis Of Variance, is used to compare the averages or means of two or more populations to better understand how they differ. Watch this tutorial for more.
Find more tutorials on the SAS Users YouTube channel.