- Home
- /
- Analytics
- /
- Stat Procs
- /
- Re: LSMEANS / LINES option in PROC GLM?
- RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
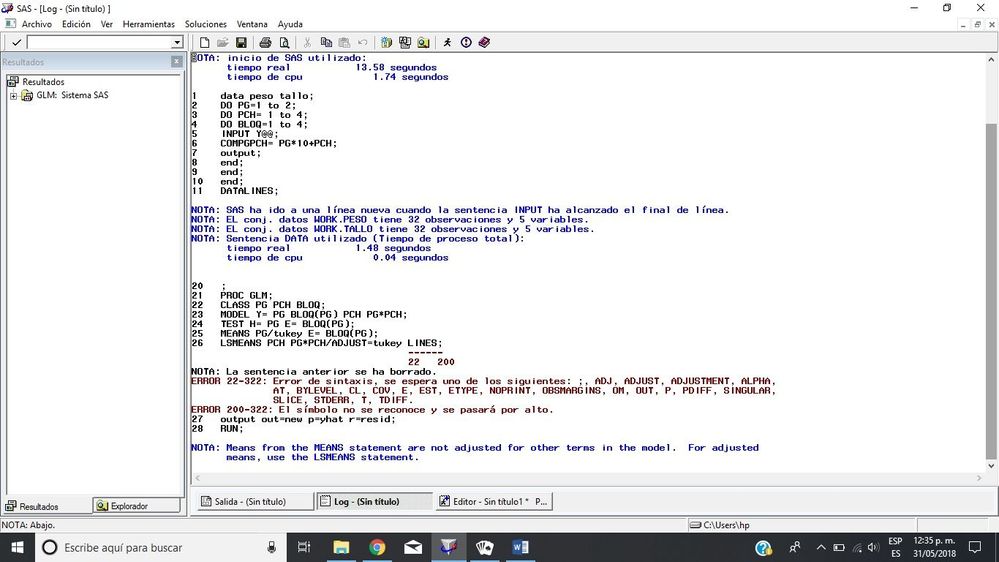
al intentar correr estos datos me salen los siguientes errores. he intentado cambiar el comando pero no salen los datos que quiero.
Accepted Solutions
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
If you have only basic SAS knowledge and you want to compare GLM to MIXED for mixed models, then you really, really need to read SAS® for Mixed Models, Second Edition.
GLM is a bad choice for mixed models because some of what it reports is incorrect. And there are good alternatives (MIXED and GLIMMIX, for example). The book I link to above covers in detail the problems that GLM has with mixed models, as well as the comparison of GLM to MIXED.
Edit: If you use your favorite search engine to look for "glm versus mixed in sas", you'll find several papers that address the comparison.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
I regret that I do not read (or speak) Spanish, so I may be answering the wrong question.
Try adding "PDIFF=ALL" as an option to the LSMEANS statement.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Thanks to try to help me. I run the command according to your observation and the error continue.
Now is a error 180-322.
I write the command that I want to analize, and the error is in the sentence "LSMEANS PCH PG*PCH/ADJUST=tukey LINES;" inclusive in lines with errors 22-200, 22-322 and 200-322.
data peso tallo;
DO PG=1 to 2;
DO PCH= 1 to 4;
DO BLOQ=1 to 4;
INPUT Y@@;
COMPGPCH= PG*10+PCH;
output;
end;
end;
end;
DATALINES;
0.91 1.13 1.18 0.88
1.52 1.36 1.87 1.31
1.07 1.18 1.26 1.65
0.68 0.62 0.81 0.70
0.81 0.72 0.79 0.74
1.23 0.90 0.90 1.07
. 1.57 1.96 1.38
0.67 0.71 0.63 0.66
;
PROC GLM;
CLASS PG PCH BLOQ;
MODEL Y= PG BLOQ(PG) PCH PG*PCH;
TEST H= PG E= BLOQ(PG);
MEANS PG/tukey E= BLOQ(PG);
LSMEANS PCH PG*PCH/ADJUST=tukey LINES;
output out=new p=yhat r=resid;
RUN;
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Your code runs without error for me, but I am using the most recent version of SAS/STAT (9.4 TS1M5). The addition of the LINES option to the GLM procedure happened in the most recent version (https://blogs.sas.com/content/iml/2017/10/16/multiple-comparisons-lines-plot.html). So if you are running an older version, that option is not available and that's why are are getting the error.
However, you are fitting a mixed model, and you'll want to switch to MIXED or GLIMMIX, which have had the LINES option for awhile. You should not use GLM to fit a mixed model, for a variety of reasons. A good resource for learning why and more is SAS® for Mixed Models, Second Edition.
Regarding your code: in your data step, you are creating two identical datasets: peso and tallo. I doubt that's your intention.
Here is an example of your model using GLIMMIX
data peso tallo;
DO PG=1 to 2;
DO PCH= 1 to 4;
DO BLOQ=1 to 4;
INPUT Y@@;
COMPGPCH= PG*10+PCH;
output;
end;
end;
end;
DATALINES;
0.91 1.13 1.18 0.88
1.52 1.36 1.87 1.31
1.07 1.18 1.26 1.65
0.68 0.62 0.81 0.70
0.81 0.72 0.79 0.74
1.23 0.90 0.90 1.07
. 1.57 1.96 1.38
0.67 0.71 0.63 0.66
;
PROC GLM;
CLASS PG PCH BLOQ;
MODEL Y= PG BLOQ(PG) PCH PG*PCH;
TEST H= PG E= BLOQ(PG);
MEANS PG/tukey E= BLOQ(PG);
LSMEANS PCH PG*PCH/ADJUST=tukey LINES;
output out=new p=yhat r=resid;
RUN;
proc glimmix data=peso;
class pg pch bloq;
model y = pg pch pg*pch;
random bloq(pg);
lsmeans pg pch / adjust=tukey lines;
lsmeans pg*pch / plot=meanplot(sliceby=pg join cl) slice=pch;
run;
I hope this helps.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
I think that the problem is my SAS´ version (SAS system for windows 9.0), I tried to run to your commmand an appear me ERROR: GLIMMIX procedure not found.
I interest to compare the pro glm and mixed, but a I confess that my knowledge in SAS are basics.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
SAS 9.0 dates back to 2002 or so--it's really outdated. If you want to use SAS, you should install the most recent version, which is probably free under the terms of your current SAS license.
Proc MIXED might do what you need, but you really ought to upgrade.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
I want to compare proc mixed split plot with glm procedure, in similar dates.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
If you have only basic SAS knowledge and you want to compare GLM to MIXED for mixed models, then you really, really need to read SAS® for Mixed Models, Second Edition.
GLM is a bad choice for mixed models because some of what it reports is incorrect. And there are good alternatives (MIXED and GLIMMIX, for example). The book I link to above covers in detail the problems that GLM has with mixed models, as well as the comparison of GLM to MIXED.
Edit: If you use your favorite search engine to look for "glm versus mixed in sas", you'll find several papers that address the comparison.
Don't miss out on SAS Innovate - Register now for the FREE Livestream!
Can't make it to Vegas? No problem! Watch our general sessions LIVE or on-demand starting April 17th. Hear from SAS execs, best-selling author Adam Grant, Hot Ones host Sean Evans, top tech journalist Kara Swisher, AI expert Cassie Kozyrkov, and the mind-blowing dance crew iLuminate! Plus, get access to over 20 breakout sessions.
ANOVA, or Analysis Of Variance, is used to compare the averages or means of two or more populations to better understand how they differ. Watch this tutorial for more.
Find more tutorials on the SAS Users YouTube channel.