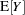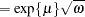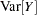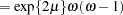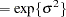- Home
- /
- Analytics
- /
- Stat Procs
- /
- Re: Issues with Proc Mixed
- RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Hi,
I am trying to run a proc mixed analysis for the following data:
ID Score year group othervariables
1 80 0 1
1 90 1 1
1 77 2 1
2 66 0 0
2 90 3 0
2 82 6 0
. .
. .
. .
So the years with scores are from 0 to 7 but some have scores years 0, 1,2, 3, 6 and some 0, 3, 6 etc. It varies. The dataset is fairly large and includes 17155 observations and 223 variables...I need to adjust for some variables in the model (around 7 etc).
When running the unadjusted model using the following code:
proc mixed data=cup.cup;
class year group;
model F393MSE = group year group*year;
repeated year/ subject=id type=ar(1);
run;
I get the following error:
WARNING: ODS graphics with more than 5000 points have been suppressed. Use the PLOTS(MAXPOINTS= )
option in the PROC MIXED statement to change or override the cutoff.
WARNING: Stopped because of infinite likelihood.
I added the PLOTS(MAXPOINTS= NONE) in the statement, but it didn't work.
Surprisingly, when I added covariates in the model (around seven) the model runs. However, I do need to run the unadjusted.
Also, one other question. How does one run a GLM with REML method and specify the random effects...
Thank you for your help!
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
You have two independent warnings. This one is only related to graphs:
WARNING: ODS graphics with more than 5000 points have been suppressed. Use the PLOTS(MAXPOINTS= )
option in the PROC MIXED statement to change or override the cutoff.
This is why the procedure stopped:
WARNING: Stopped because of infinite likelihood.
Changing the maximum number of points plotted will not affect the likelihood. Changing the model will.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Try repeated year/ subject=id type=chol ;
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Thank you ksharp. I tried repeated year/ subject=id type=chol;
and I don't think chol is an option for the variance covariance structure. I got the following error:
ERROR 22-322: Syntax error, expecting one of the following: ANTE, AR, ARH, ARMA, CS, CSH, FA,
FA0, FA1, HF, LIN, LINEAR, SIMPLE, SP, TOEP, TOEPH, UN, UNAR, UNCS, UNR, VC.
any additional ideas? Is it possible for me to try proc glm with REML?
Thanks.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
If you have a mixed design, the MIXED (or GLIMMIX) procedure is much better than GLM; in fact, you generally do not want to use the GLM procedure with a mixed model.
Is the dataset sorted by ID? In the current model ID is a continuous variable, and because you are using it as a SUBJECT in the REPEATED statement, the dataset has to be sorted by ID.
What are the distributional properties of F393MSE? Is this variable measured on a continuous scale? Is normality a reasonable assumption?
Would it make any sense to regress on year?
Do you still get an error:
1) if you add ID to the CLASS statement
2) if you use
repeated year/ subject=id type=cs;
If none of these provide any insight into your problem, then probably you'll need to provide data.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Thank you sld...
Sorry for the delay in sending an update...
The model worked when transforming the outcome which is a continuous scale to a logarithmic scale.
However, what are the options in running the model on the logarithmic scale and then generating the lsmeans and plot on the regular scale?
Thanks..
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
The ILINK option does not function for DIST=LOGNORMAL. To get what you want, you can re-transform estimates produced (on the log scale) by the model "by hand"--either literally by hand, or save estimates to a SAS dataset and do the conversion in a data step or even in Excel.
See the GLIMMIX > MODEL > DIST= documentation http://documentation.sas.com/?docsetId=statug&docsetVersion=14.2&docsetTarget=statug_glimmix_syntax1...
which says:
When you choose DIST=LOGNORMAL, the GLIMMIX procedure models the logarithm of the response variable as a normal random variable. That is, the mean and variance are estimated on the logarithmic scale, assuming a normal distribution,
. This enables you to draw on options that require a distribution in the exponential family—for example, by using a scoring algorithm in a GLM. To convert means and variances for
into those of
, use the relationships
Also see this thread https://communities.sas.com/t5/General-SAS-Programming/ilink-will-not-return-inverse-values-in-the-l...
HTH 🙂
Don't miss out on SAS Innovate - Register now for the FREE Livestream!
Can't make it to Vegas? No problem! Watch our general sessions LIVE or on-demand starting April 17th. Hear from SAS execs, best-selling author Adam Grant, Hot Ones host Sean Evans, top tech journalist Kara Swisher, AI expert Cassie Kozyrkov, and the mind-blowing dance crew iLuminate! Plus, get access to over 20 breakout sessions.
ANOVA, or Analysis Of Variance, is used to compare the averages or means of two or more populations to better understand how they differ. Watch this tutorial for more.
Find more tutorials on the SAS Users YouTube channel.