- Home
- /
- Analytics
- /
- Stat Procs
- /
- Help with multiple comparisons test in Proc Lifetest (SAS 9.4)
- RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
I dispose of a dataset on kidney transplant patients and I am looking at the survival time difference between several kidney diseases after transplantation.
Summary of the data:
- group 1: 66 patients, 20 events
- group 2: 83 patients, 8 events
- group 3: 702 patients, 53 events
Non-events are being right-censored.
After running the following 'proc lifetest', we end up with this survival plot:
proc lifetest data=DATASET plots=survival;
time time*Death(0);
strata disease / adjust=tukey;
run;
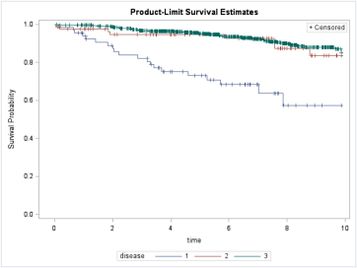
We found a significant (p<0.0001) Log-Rank test and significant post-hoc comparisons between all the groups. So, in contrast to what the figure suggests, we found a significant difference between disease 2 and 3 (p=0.0257 after Tukey adjustement).
I ran the same analysis in R with the package survminer and found no significant difference between the two groups. In fact, it appeared that the post-hoc testing in R is based on the Log-Rank test including only the groups of interest. And indeed, if we would run a proc lifetest on a dataset including only disease 2 and 3, the same, non-significant p-value (p=0.58) was found.
After inspecting the SAS algoritjm, explained in: https://support.sas.com/documentation/cdl/en/statug/63033/HTML/default/viewer.htm#statug_lifetest_a0...
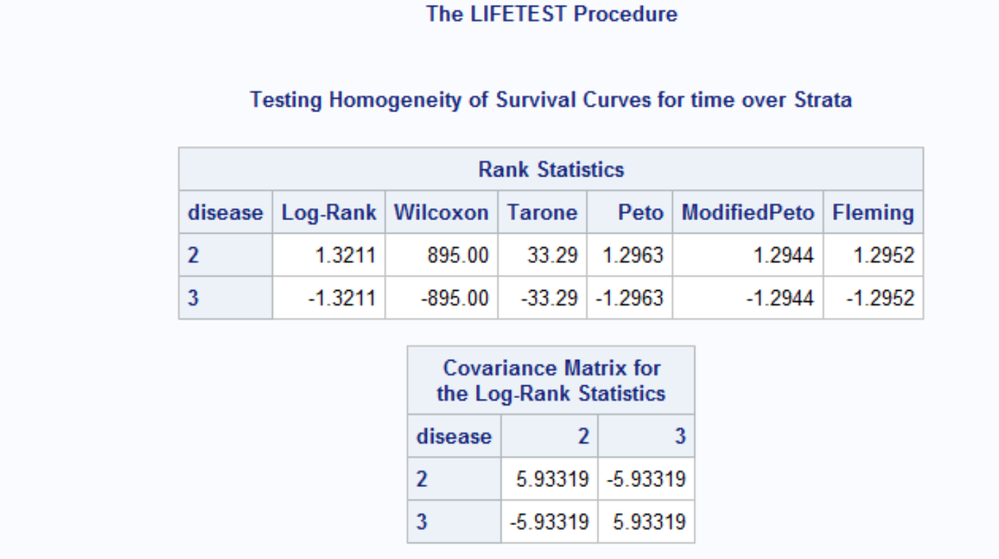
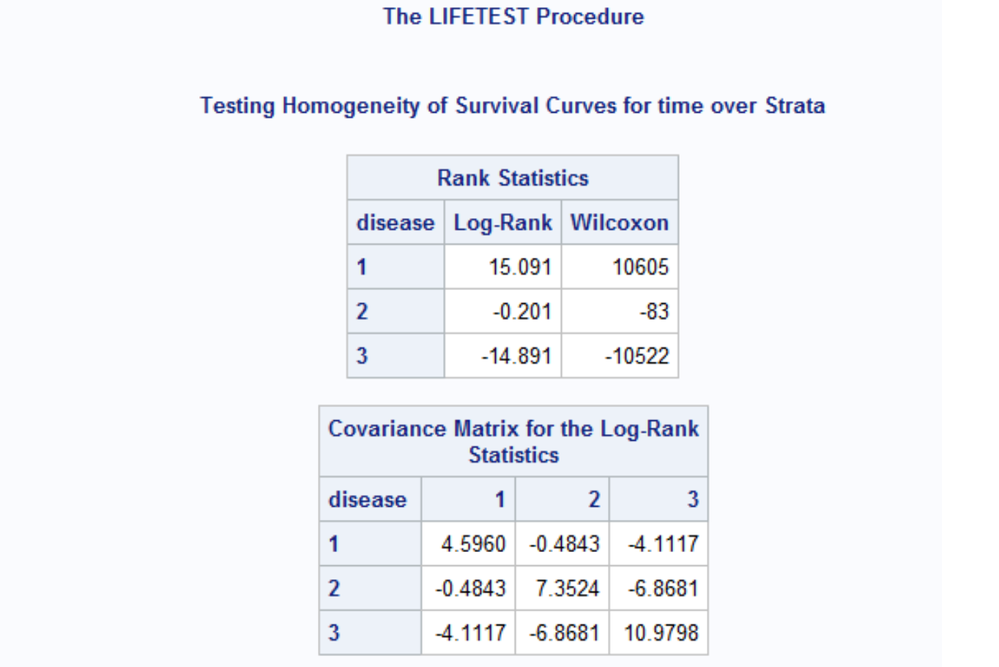
we saw that the multiple comparisons test statistic 'z²jl' includes data on the pooled sample. So, when comparing diseases 2 versus 3, data on disease 1 is implicitly involved in the algorithm. This is reflected in the difference between the 'Rank Statistics' and their 'Covariance matrix'. See:
- log-rank statistic and covariance matrix using 2 groups only
- log-rank statistic and covariance matrix using 3 groups
Let's say this kind of post-hoc Log-Rank testing is based on the rationale of post-hoc testing in ANOVA, where it is possible that a post-hoc test provides different results than the separate t-tests. However, in our case the p-values differ hugely and, above all, it is rather difficult to argue that disease 2 and 3 show a significantly different survival based on the KM-plot shown earlier.
I noticed that large parts of the SAS documentation refer to the work of Klein and Moeschberger, 1997. Yet, when inspecting this work, very little is being said about multiple testing. The only relevant remarks I could deduce were:
(p.237) "If one is interested in comparing K groups in a pairwise simultaneous manner then an adjustment for multiple tests must be made. One such method that can be used is the Bonferroni method of multiple comparisons."
(p.241) "Using the log-rank test, perform the three pairwise tests of the hypothesis [...] For each test, use only those individuals with stage j or j +1 of the disease. Make an adjustment to your critical value for multiple testing to give an approximate 0.05 level test."
Also, I have found no literature on a post-hoc Log-Rank test statistic that involves using the pooled sample.
In 2012 a similar discussion was started on this forum:
The answer that the statistical significance is caused by the sample size is not really satisfying to me. I know my sample size are varying greatly, but I don't believe this is the problem.
The larger issue for me, is that there seems to be no consistency across different tests and that SAS makes use of a test statistic of which I cannot find any documentation.
Can anyone provide me with some insight into this matter?
Thanks,
Maarten
Accepted Solutions
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
And there it is:
Dear Dhr. Maarten Coemans,
Our R&D team concluded the following:
While the multiple comparisons procedure in LIFETEST does tend to have inflated type 1 errors when the groups are highly unbalanced (as your example), the performance is the expected under more balanced settings. As an alternative, we suggest using PROC MULTTEST to perform post hoc multiple comparisons adjustment to the pairwise (unadjusted) p-values that LIFETEST produce. This way, the adjustment would be purely on the p-values and does not involve the global log-rank statistic.
In the future, we may consider adding the method proposed in the Statistics in Medicine paper by Logan et al. as another alternative.
We hope this information is helpful. Please let us know if additional clarification is needed..
Thank you
Kind Regards
The article they are referring to is: 'Pairwise multiple comparison adjustment in survival analysis'.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
@MaartenC Have you got any updates on this? I am working on a similar project, and is thinking about the same issue with justifying multiple comparison issue.
However as you mentioned, if we use Tukey, since it is a method that adjusts for multiple comparison, it does use the pooled sample (which means in the Tukey method,whenever you are looking at two groups, the information of other groups will be involved).
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
I had contact with the SAS technical support about half a year ago. They said they were going run some simulations to check Type I error rates, etc... Haven't heard from them since, but send an email just now for an update on this issue.
In my opinion, this procedure should not be used and you better perform separate pairwise comparisons, with a possible adjustment to the p-values afterwards.
I'll keep you posted about their answer.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
And there it is:
Dear Dhr. Maarten Coemans,
Our R&D team concluded the following:
While the multiple comparisons procedure in LIFETEST does tend to have inflated type 1 errors when the groups are highly unbalanced (as your example), the performance is the expected under more balanced settings. As an alternative, we suggest using PROC MULTTEST to perform post hoc multiple comparisons adjustment to the pairwise (unadjusted) p-values that LIFETEST produce. This way, the adjustment would be purely on the p-values and does not involve the global log-rank statistic.
In the future, we may consider adding the method proposed in the Statistics in Medicine paper by Logan et al. as another alternative.
We hope this information is helpful. Please let us know if additional clarification is needed..
Thank you
Kind Regards
The article they are referring to is: 'Pairwise multiple comparison adjustment in survival analysis'.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Thanks a lot. Really appreciate the feedback.
Don't miss out on SAS Innovate - Register now for the FREE Livestream!
Can't make it to Vegas? No problem! Watch our general sessions LIVE or on-demand starting April 17th. Hear from SAS execs, best-selling author Adam Grant, Hot Ones host Sean Evans, top tech journalist Kara Swisher, AI expert Cassie Kozyrkov, and the mind-blowing dance crew iLuminate! Plus, get access to over 20 breakout sessions.
ANOVA, or Analysis Of Variance, is used to compare the averages or means of two or more populations to better understand how they differ. Watch this tutorial for more.
Find more tutorials on the SAS Users YouTube channel.