- Home
- /
- Programming
- /
- Graphics
- /
- How to remove the green observation's correlation loading points in co...
- RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Hi All,
I used Proc PLS to do a multivariate analysis and got a plot as attached. How can I remove the green colored points in the picture? I think they are the observations' correlation values. For example, I have 90 observations, and each of them will have a loading value on factor1 and factor2, so there will be 90 green points shown in the picture. Who can tell me which option can suppress them? for example, data is like this:
par1 par2 par3 par4 par5 par6 par7 location
2680 0.546089996 237 1 0.172 2.25 305 5
3750 0.54836587 140 1.55 0.111 1.06 425 5
3590 0.54878718 168 1.27 0.131 0.969 516 5
2390 0.549510935 183 1.07 0.096 1.84 260 5
3780 0.549631747 140 1.12 0.118 1.98 472 5
2790 0.549934008 200 1.1 0.221 2.13 321 5
2880 0.5499945 227 1.14 0.185 1.54 439 5
2910 0.550357733 259 1.31 0.116 1.31 289 5
2420 0.550842789 177 1.32 0.044067423 1.95 260 5
3850 0.550964187 128 1.41 0.117 1.08 471 5
3530 0.552425146 165 1.23 0.11 1.57 494 5
2730 0.552913856 223 1.03 0.17 2 330 5
3130 0.553158535 252 1.02 0.174 2.13 322 5
3040 0.553709856 272 1.21 0.155 1.97 317 5
3830 0.554139421 153 1.27 0.137 1.47 455 5
3930 0.554569654 164 1.17 0.116 1.5 481 5
2430 0.554569654 136 1.3 0.198 2.11 226 8
3630 0.555247085 137 1.17 0.1 1.75 413 5
2490 0.555432126 176 1.06 0.113 1.39 236 5
3490 0.555555556 166 1.28 0.044444444 1.65 465 5
3840 0.556173526 164 1.23 0.0949 1.66 470 5
2480 0.556173526 239 1.28 0.102 2.2 238 5
3760 0.556173526 191 1.33 0.131 2.12 447 5
3850 0.556173526 174 1.35 0.241 2.42 381 3
3410 0.557413601 174 1.14 0.107 1.48 419 5
2960 0.559284116 229 1.08 0.165 1.99 304 5
3410 0.559284116 137 1.19 0.291 2.17 375 8
3300 0.560538117 121 1.13 0.153 1.82 352 8
3090 0.560538117 134 1.16 0.167 1.17 416 4
3210 0.560538117 124 1.09 0.172 0.82 390 4
3950 0.560538117 130 1.29 0.199 1.89 440 4
3300 0.561167228 131 1.06 0.242 2.45 367 8
2210 0.561167228 162 0.885 0.288 3.32 208 4
3170 0.561797753 126 1.3 0.151 1.31 388 4
2740 0.561797753 96.1 1.22 0.245 0.827 254 3
3750 0.561797753 144 1.08 0.257 2.62 366 3
3640 0.562429696 120 1.32 0.159 1.63 347 8
3210 0.563063063 148 1.29 0.206 2.18 352 8
2300 0.563697858 179 0.936 0.181 2.29 223 2
3410 0.564334086 141 0.856 0.136 2.03 370 8
3500 0.564334086 126 1.38 0.177 1.45 355 8
3470 0.564334086 101 0.989 0.222 1.84 349 3
2260 0.564334086 171 0.942 0.224 2.08 219 2
2220 0.564334086 180 0.956 0.281 1.84 219 4
2340 0.564971751 165 1.05 0.228 2.25 240 8
2380 0.564971751 161 0.976 0.287 1.6 214 4
3220 0.56561086 148 1.21 0.121 0.568 520 6
3920 0.566251416 176 1.08 0.045300113 2.26 637 6
3830 0.566251416 137 1.48 0.203 1.23 387 3
2510 0.566251416 152 1.24 0.222 1.84 223 8
2760 0.566251416 168 0.994 0.282 1.31 280 4
2640 0.566251416 154 0.979 0.345 1.52 291 4
3570 0.566893424 165 1.33 0.155 2.18 505 6
3170 0.566893424 126 1.08 0.162 1.41 341 4
3700 0.566893424 159 1.3 0.17 1.64 449 4
3250 0.566893424 104 1.32 0.2 1.37 372 8
3740 0.566893424 159 1.23 0.216 1.69 409 1
3380 0.566893424 163 1.53 0.245 2.19 367 3
3240 0.56753689 136 1.07 0.153 1.88 383 4
3400 0.56753689 109 1.36 0.161 1.16 420 4
3760 0.56753689 150 0.93 0.169 1.68 537 4
3560 0.56753689 123 1.03 0.193 2.32 374 8
2360 0.56753689 163 0.697 0.235 1.94 243 8
2430 0.56753689 166 0.762 0.247 2.31 231 8
3330 0.568181818 148 1.11 0.174 2 393 4
3080 0.568181818 139 1.13 0.188 2.08 349 8
3230 0.568181818 116 1.23 0.199 1.77 328 8
2180 0.568181818 144 1.01 0.215 2.13 207 8
2520 0.568181818 128 0.809 0.369 1.65 306 4
3320 0.568828214 152 1.15 0.14 1.65 395 4
2300 0.568828214 134 0.908 0.221 1.56 233 8
3730 0.568828214 141 1.58 0.238 1.96 405 3
3800 0.568828214 160 1.24 0.241 2.2 402 3
2440 0.568828214 153 1.03 0.258 1.89 223 4
3910 0.568828214 209 1.26 0.275 2.26 350 3
4010 0.569476082 139 1.28 0.045558087 1.7 602 6
2340 0.570125428 167 1.1 0.18 1.57 208 2
2360 0.570125428 176 0.704 0.2 1.6 219 2
3490 0.570776256 171 1.43 0.269 2.4 360 3
2620 0.571428571 132 1.09 0.202 1.8 224 8
3740 0.571428571 172 1.27 0.256 1.92 355 3
3600 0.57208238 128 1.16 0.17 1.94 434 4
3360 0.57208238 150 1.18 0.171 1.81 353 1
3620 0.57208238 131 1.28 0.177 2.24 360 3
3560 0.57208238 139 1.15 0.229 1.9 366 3
2740 0.572737686 277 0.876 0.171 1.71 290 10
2340 0.572737686 148 0.964 0.231 1.18 250 6
2760 0.572737686 168 0.905 0.303 2.1 264 4
2890 0.572737686 204 0.857 0.331 2.32 272 2
code is :
proc pls data=check method=rrr;
class location;
model par1-par7=location;
run;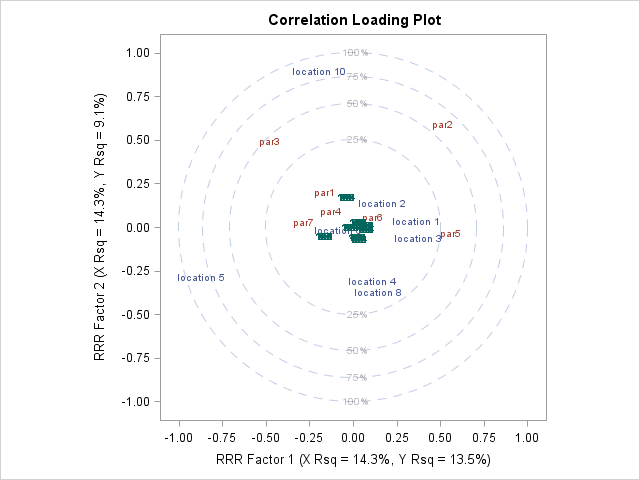
Accepted Solutions
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
See if this helps. I looked at the data object and graph template then I eliminated (more precisely set to missing) the CORRX and CORRY observations when CORRGROUP was 'Observation' You can do template modifications in an editor, but I prefer to use a DATA step in order to provide reproducible results.
proc template;
delete Stat.PLS.Graphics.CorrLoadPlot;
source Stat.PLS.Graphics.CorrLoadPlot / file='temp.temp';
quit;
data _null_;
infile 'temp.temp';
input;
if index(_infile_, 'CORRGROUP') then do;
_infile_ = tranwrd(_infile_, 'x=CORRX ',
"x=eval(ifn(corrgroup = 'Observation', ., corrx)) ");
_infile_ = tranwrd(_infile_, 'y=CORRY ',
"y=eval(ifn(corrgroup = 'Observation', ., corry)) ");
end;
if _n_ = 1 then call execute('proc template;');
call execute(_infile_);
run;
proc pls data=check method=rrr;
class location;
model par1-par7=location;
run;
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
See if this helps. I looked at the data object and graph template then I eliminated (more precisely set to missing) the CORRX and CORRY observations when CORRGROUP was 'Observation' You can do template modifications in an editor, but I prefer to use a DATA step in order to provide reproducible results.
proc template;
delete Stat.PLS.Graphics.CorrLoadPlot;
source Stat.PLS.Graphics.CorrLoadPlot / file='temp.temp';
quit;
data _null_;
infile 'temp.temp';
input;
if index(_infile_, 'CORRGROUP') then do;
_infile_ = tranwrd(_infile_, 'x=CORRX ',
"x=eval(ifn(corrgroup = 'Observation', ., corrx)) ");
_infile_ = tranwrd(_infile_, 'y=CORRY ',
"y=eval(ifn(corrgroup = 'Observation', ., corry)) ");
end;
if _n_ = 1 then call execute('proc template;');
call execute(_infile_);
run;
proc pls data=check method=rrr;
class location;
model par1-par7=location;
run;
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
##- Please type your reply above this line. Simple formatting, no
attachments. -##
Don't miss out on SAS Innovate - Register now for the FREE Livestream!
Can't make it to Vegas? No problem! Watch our general sessions LIVE or on-demand starting April 17th. Hear from SAS execs, best-selling author Adam Grant, Hot Ones host Sean Evans, top tech journalist Kara Swisher, AI expert Cassie Kozyrkov, and the mind-blowing dance crew iLuminate! Plus, get access to over 20 breakout sessions.
Learn how use the CAT functions in SAS to join values from multiple variables into a single value.
Find more tutorials on the SAS Users YouTube channel.
 Click image to register for webinar
Click image to register for webinar
Classroom Training Available!
Select SAS Training centers are offering in-person courses. View upcoming courses for:



