- Home
- /
- Programming
- /
- Programming
- /
- Split header on top of Spanning Header in proc report
- RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Hello,
I would like to create a split header on top of a spanning header, but having trouble figuring out how to organize the code.
Here is what I have:
proc report data=all&n out=vall&n (where=(_break_='' and paramn ^=.))
headline headskip nowindows missing split = "@"
style=[protectspecialchars=off] style(report)=[bordertopwidth=1.5];
columns sp parcat2n paramn param
("(*ESC*)S={borderbottomcolor=black borderbottomwidth=2} Treatment A@(N=&N0.)"
_0LOW _0HIGH)
("(*ESC*)S={borderbottomcolor=black borderbottomwidth=2} Treatment B@(N=&N1.)"
_1LOW _1HIGH)
("(*ESC*)S={borderbottomcolor=black borderbottomwidth=2} Overall Post-Baseline"
_0LOW _0HIGH _1LOW _1HIGH);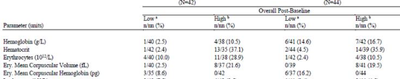
Accepted Solutions
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Hi:
What you currently show is what PROC REPORT will give you. I do not think there's a way to get PROC REPORT to do what you want to do.
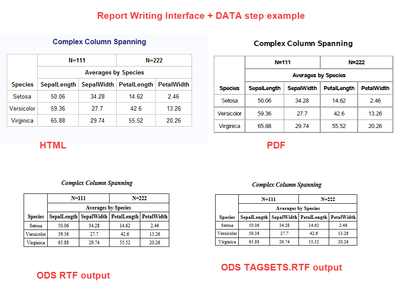
I was pleasantly surprised when testing the Report Writing Interface that my program generated the correct results in both ODS RTF and ODS TAGSETS.RTF (yay!), as shown below:
I just used SASHELP.IRIS, and presummarized it to get the averages. There are 4 columns in the report plus the Species column, to mimic your columns and then using some headers that are similar to what you want, just as a proof of concept. Here's the code I used:
%let mypath=c:\temp;
** presummarize IRIS data to get only averages;
proc means data=sashelp.iris mean maxdec=2;
class species;
var sepallength sepalwidth petallength petalwidth;
output out=work.allavg mean=;
run;
ods html path="&mypath." file="complex_column_span.html";
ods pdf file="&mypath.\complex_column_span.pdf" ;
ods rtf file="&mypath.\odsr_column_span.rtf";
ods tagsets.rtf file="&mypath.\odstr_column_span.rtf";
title "Complex Column Spanning";
data _null_;
set allavg(keep=_type_ species sepallength sepalwidth petallength petalwidth)
end=last;
by species;
if _N_ = 1 then do;
dcl odsout obj();
obj.table_start();
obj.head_start();
** Header row 1;
obj.row_start(type:"Header");
obj.format_cell(text: "Species", row_span:3,style_attr:"color=black backgroundcolor=white vjust=b fontweight=bold");
obj.format_cell(text: "N=111", column_span:2, style_attr:"color=black backgroundcolor=white");
obj.format_cell(text: "N=222", column_span:2, style_attr:"color=black backgroundcolor=white");
obj.row_end();
** Header row 2;
obj.row_start(type:"Header");
obj.format_cell(text: "Averages by Species", column_span:4, style_attr:"color=black backgroundcolor=white");
obj.row_end();
** Header row 3;
obj.row_start(type: "Header");
obj.format_cell(text: "SepalLength", style_attr:"color=black backgroundcolor=white fontweight=bold");
obj.format_cell(text: "SepalWidth", style_attr:"color=black backgroundcolor=white fontweight=bold");
obj.format_cell(text: "PetalLength", style_attr:"color=black backgroundcolor=white fontweight=bold");
obj.format_cell(text: "PetalWidth", style_attr:"color=black backgroundcolor=white fontweight=bold");
obj.row_end();
obj.head_end();
end;
** row for every obs;
if _type_ = 1 then do;
obj.row_start();
obj.format_cell(data: species, row_span:1);
obj.format_cell(data: sepallength, row_span:1);
obj.format_cell(data: sepalwidth, row_span:1);
obj.format_cell(data: petallength, row_span:1);
obj.format_cell(data: petalwidth, row_span:1);
obj.row_end();
end;
if last then do;
obj.table_end();
end;
run;
ods html close;
ods pdf close;
ods rtf close;
ods tagsets.rtf close;
title; footnote;
I used the _TYPE_=1 rows from the PROC MEANS OUTPUT data set because I did not need the _TYPE_=0 row.
Depending on the structure of your data and what you need to do, the Report Writing Interface may be an option.
Cynthia
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Without any data or the full code, nobody can play with your example because they either have to guess at the structure of your data or they have to guess what the rest of your program is. You could be trying to get either HTML, RTF or PDF output, so knowing your ODS statements would be useful too. In just a quick look, it's going to be hard to split the column headers the way you show using PROC REPORT. Spanning headers such as you show are more like the type of thing you can do with the Report Writing Interface (RWI) and the DATA step. However, that brings us back to what destination you want because the RWI Interface is not supported in all destinations.
Cynthia
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Are you actually creating multiple columns with the same values?
Removing the header text that columns statement reduces to
columns sp parcat2n paramn param _0LOW _0HIGH _1LOW _1HIGH _0LOW _0HIGH _1LOW _1HIGH;
Which has 4 variables repeated and seems like an odd thing to do.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
ods listing close;
ods tagsets.rtf file="&outpath" options(vspace='NO') &repopt;
title3 "&ttlnum";
title4 "&ttldesc";
title5 "&ttlpop";
title6 " ";
proc report data=all&n out=vall&n (where=(_break_='' and paramn ^=.))
headline headskip nowindows missing split = "@"
style=[protectspecialchars=off] style(report)=[bordertopwidth=1.5];
columns sp parcat2n paramn param
("(*ESC*)S={borderbottomcolor=black borderbottomwidth=2} Overall Post-Baseline"
("(*ESC*)S={borderbottomcolor=black borderbottomwidth=2} Treatment A@(N=&N0.)"
_0LOW _0HIGH)
("(*ESC*)S={borderbottomcolor=black borderbottomwidth=2} Treatment B@(N=&N1.)"
_1LOW _1HIGH));
define sp /order order=internal noprint;
define parcat2n /order order=internal noprint;
define paramn /order order=internal noprint;
define param / display style=[cellwidth=2.85 in just=l asis=on] "Parameter (units)";
define _0LOW / display style=[cellwidth=1.5 in just=c asis=on] "Low ~{super a}@n/nn (%)";
define _0HIGH / display style=[cellwidth=1.5 in just=c asis=on] "High ~{super b}@n/nn (%)";
define _1LOW / display style=[cellwidth=1.5 in just=c asis=on] "Low ~{super a}@n/nn (%)";
define _1HIGH / display style=[cellwidth=1.5 in just=c asis=on] "High ~{super b}@n/nn (%)";
break after parcat2n / page;
compute before sp;
line put ' ';
endcomp;
run;
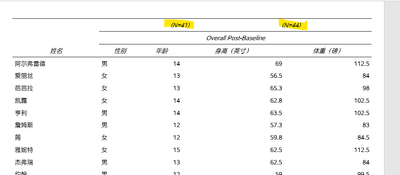
Sorry not duplicating the columns ( I was playing around with the code earlier and realized I didn't revert back). This current code spans 'overall post-baseline' at top, but I actually need it below the split header (Treatment A and Treatment B).
Currently:
Need:
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Not a direct solution of inserting a single spanning UNDER two (or more) other headings but for some appearances may give a similar enough appearance:
proc report data=sashelp.class headline headskip;
columns name ('header first columns'
(" "
sex age))
(' next block'
("other secondline header"
height weight))
;
run;
Not trying to deal style appearance options as more of demonstration of concept.
With horizontal justification options in the first line, second and actual variable headers then the "other secondline header" , while not exactly centered (assumption of desire) may be close enough to fool the eye.
Specifying exact column widths for the variable columns would likely help as well.
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Hi:
What you currently show is what PROC REPORT will give you. I do not think there's a way to get PROC REPORT to do what you want to do.
I was pleasantly surprised when testing the Report Writing Interface that my program generated the correct results in both ODS RTF and ODS TAGSETS.RTF (yay!), as shown below:
I just used SASHELP.IRIS, and presummarized it to get the averages. There are 4 columns in the report plus the Species column, to mimic your columns and then using some headers that are similar to what you want, just as a proof of concept. Here's the code I used:
%let mypath=c:\temp;
** presummarize IRIS data to get only averages;
proc means data=sashelp.iris mean maxdec=2;
class species;
var sepallength sepalwidth petallength petalwidth;
output out=work.allavg mean=;
run;
ods html path="&mypath." file="complex_column_span.html";
ods pdf file="&mypath.\complex_column_span.pdf" ;
ods rtf file="&mypath.\odsr_column_span.rtf";
ods tagsets.rtf file="&mypath.\odstr_column_span.rtf";
title "Complex Column Spanning";
data _null_;
set allavg(keep=_type_ species sepallength sepalwidth petallength petalwidth)
end=last;
by species;
if _N_ = 1 then do;
dcl odsout obj();
obj.table_start();
obj.head_start();
** Header row 1;
obj.row_start(type:"Header");
obj.format_cell(text: "Species", row_span:3,style_attr:"color=black backgroundcolor=white vjust=b fontweight=bold");
obj.format_cell(text: "N=111", column_span:2, style_attr:"color=black backgroundcolor=white");
obj.format_cell(text: "N=222", column_span:2, style_attr:"color=black backgroundcolor=white");
obj.row_end();
** Header row 2;
obj.row_start(type:"Header");
obj.format_cell(text: "Averages by Species", column_span:4, style_attr:"color=black backgroundcolor=white");
obj.row_end();
** Header row 3;
obj.row_start(type: "Header");
obj.format_cell(text: "SepalLength", style_attr:"color=black backgroundcolor=white fontweight=bold");
obj.format_cell(text: "SepalWidth", style_attr:"color=black backgroundcolor=white fontweight=bold");
obj.format_cell(text: "PetalLength", style_attr:"color=black backgroundcolor=white fontweight=bold");
obj.format_cell(text: "PetalWidth", style_attr:"color=black backgroundcolor=white fontweight=bold");
obj.row_end();
obj.head_end();
end;
** row for every obs;
if _type_ = 1 then do;
obj.row_start();
obj.format_cell(data: species, row_span:1);
obj.format_cell(data: sepallength, row_span:1);
obj.format_cell(data: sepalwidth, row_span:1);
obj.format_cell(data: petallength, row_span:1);
obj.format_cell(data: petalwidth, row_span:1);
obj.row_end();
end;
if last then do;
obj.table_end();
end;
run;
ods html close;
ods pdf close;
ods rtf close;
ods tagsets.rtf close;
title; footnote;
I used the _TYPE_=1 rows from the PROC MEANS OUTPUT data set because I did not need the _TYPE_=0 row.
Depending on the structure of your data and what you need to do, the Report Writing Interface may be an option.
Cynthia
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
If you are using JOURNAL style,you could pad some white blanks to mimic this kind of style .
options nonumber nodate;
ods rtf file='c:\temp\temp.rtf' style=journal;
title;
ods escapechar='~';
proc report data=sashelp.class nowd style={outputwidth=100%};
columns name ('~S={borderbottomcolor=black borderbottomwidth=2}(N=41) (N=44)'
('~S={borderbottomcolor=black borderbottomwidth=2}Overall Post-Baseline' sex age height weight))
;
run;
ods rtf close;
- Mark as New
- Bookmark
- Subscribe
- Mute
- RSS Feed
- Permalink
- Report Inappropriate Content
Available on demand!
Missed SAS Innovate Las Vegas? Watch all the action for free! View the keynotes, general sessions and 22 breakouts on demand.
Learn how use the CAT functions in SAS to join values from multiple variables into a single value.
Find more tutorials on the SAS Users YouTube channel.
 Click image to register for webinar
Click image to register for webinar
Classroom Training Available!
Select SAS Training centers are offering in-person courses. View upcoming courses for: